Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein synthesis in artificial cells: using compartmentalisation for spatial organisation in vesicle bioreactors†
Yuval
Elani
ab,
Robert V.
Law
ab and
Oscar
Ces
*ab
aDepartment of Chemistry, Imperial College London, Exhibition Road, SW7 2AZ, UK. E-mail: o.ces@imperial.ac.uk
bInstitute of Chemical Biology, Imperial College London, Exhibition Road, SW7 2AZ, UK
First published on 1st May 2015
Abstract
Whereas spatial organisation of function is ubiquitous in biology, it has been lacking in artificial cells. We rectify this by using multi-compartment vesicles as chassis for artificial cells, allowing distinct biological processes to be isolated in space. This is demonstrated by in vitro synthesis of two proteins in predefined vesicle regions.
Artificial cells are constructs that have been built from the bottom up using biological or synthetic building blocks, which mimic biological cells in morphology or function. In bottom-up synthetic biology lipid vesicles are widely used as cellular models and as chassis for artificial cells. They have been used to host a range of biological and biomimetic processes, including reproduction,1 biomolecule synthesis,2 cell–cell communication,3,4 and sense/response behaviours.5 They have been utilized as mimics of multicellular assemblies6–8 and as simplified models of bio-membranes, allowing insights into the potential role of membrane biophysics in modulating cellular processes.9–11 Vesicles are considered an attractive platform for such applications due to their biological compatibility, their ability to host transmembrane proteins, their rich phase behaviour, and their resemblance of cellular membranes.
One of the defining characteristics of biological cells is their spatial organisation of content and function, with different biochemical processes confined to specific cellular regions. In contrast, artificial cells have had largely homogenous interiors. In order for artificial cells to start mimicking the complexity of their biological counterparts, and for them to exhibit higher order properties, this divide needs to be bridged. Herein we use compartmentalisation as an engineering principle to rectify this: multi-compartment vesicles are used as chassis for artificial cells that have specific regions devoted to different biological processes.
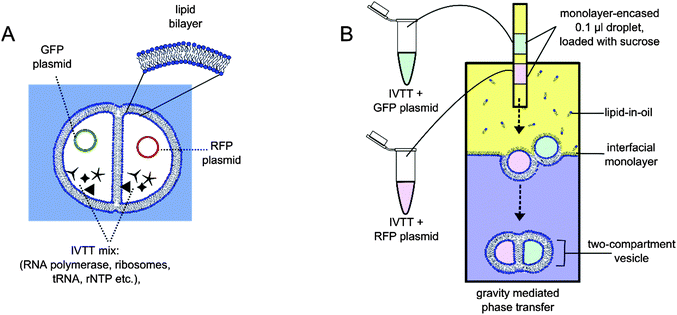
We have previously developed methods to construct vesicles with defined compartment number, content and size, where compartments were separated from one another via a lipid bilayer.12 We also used such multi-compartment vesicles as microreactors that hosted a multi-step reaction cascade, mimicking a cellular signalling pathway.13 This work builds on this: we demonstrate protein synthesis using in vitro transcription and translation (IVTT), using the separation of content to achieve segregation of function. We show for the first time that each compartment can be devoted to the biochemical synthesis of distinct biomolecules—green fluorescent protein (GFP) being synthesised in one compartment, red fluorescent protein (RFP) in another (Fig. 1). In doing so we succeed in using compartmentalisation to introduce a level of spatial organisation of function that has not been previously seen in artificial cells.
For cell-free protein expression we used the PURExpress14 IVTT kit that contained the biochemical components necessary to synthesise proteins in the presence of a plasmid. The reaction mix was made by adding the kit components to plasmids encoding for fluorescent proteins, as well as an added volume to yield a final solution composition of 0.9 M sucrose and 9 mM 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) liposomes (see ESI† for experimental details). Two such solutions were made: one containing Dasher-GFP plasmid, the other an RFP plasmid.
Two-compartment vesicles were generated according to previously published methods, using the phase transfer of two water-in-oil droplets (Fig. 1 and ESI† for details).12,13 Briefly, a water/oil column was first made, with a 4.7 mM DPhPC lipid-in-oil solution. This was left to stabilise for two hours, resulting in a densely packed monolayer at the water/oil interface. Two sucrose-loaded lipid-stabilised droplets were then expelled above the same location of the column: one droplet containing the GFP plasmid, the other the RFP plasmid. As these droplets descended through the column under gravity, they made contact with another resulting in a droplet interface bilayer. As the droplet pair entered the aqueous phase of the column, they were enveloped by a second monolayer, encasing both droplets together in a bilayer, and resulting in dual-compartment vesicles with defined content in each compartment. Immediately after vesicles were generated, 1.5 M sucrose was injected into the bottom of the aqueous phase to reduce the osmotic imbalance between the vesicle interior and exterior.
Protein synthesis was followed over time by monitoring fluorescence using a FITC filter for GFP and a TRITC filter for RFP at 100 ms and 1000 ms exposures respectively (Fig. 2). Over the course of 50 minutes at 37 °C, fluorescence of both compartments was observed to increase concurrently, indicating successful spatially segregated protein synthesis: different regions of the vesicle were specialised to perform distinct biological processes, acting as modules within the artificial cell system. Furthermore, as biological macromolecules are largely impermeable to membranes, the process remained spatially isolated over the course of the experiment.
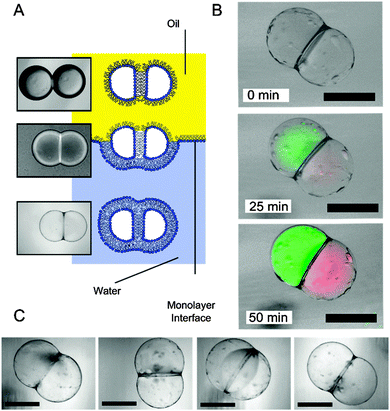 | ||
| Fig. 2 (A) Schematic and brightfield images showing the transformation from a droplet pair in an oil environment to a two-compartment vesicle surrounded by a bulk aqueous solution. Note the contrast difference in the three different stages. (B) Brightfield–fluorescent composite images of a spatially segregated artificial cell at different times after they were generated. Cell-free protein synthesis from template DNA was successful, with GFP (green channel) synthesised in one region, RFP (red channel) in another. (C) Brightfield images of several two-compartment vesicles that were generated. As previously noted, oil pockets were seen to accumulate on the vesicle surface.12 Scale bar = 200 µm. | ||
Vesicles were found to be formed with a lower efficiency than those previously reported, with approximately 12% successful rate of formation (n = 50). This was attributed to the presence of a relatively large concentration of macromolecules (estimated at 2.6 mg ml−1 by the supplier), which are thought to interfere with the formation of a well-packed monolayer at the water/oil interface of the droplet precursors. Friddin et al. observed the same effect in a droplet interface bilayer system and specifically attributed it to the energy supply fraction of the IVTT kit,15 which destabilised the interface. In a similar manner to our experiments, they (and others)16 counteracting this by adding liposomes inside the droplets to increase the probability of sufficient monolayer coverage.
On the occasions where phase transfer was unsuccessful, vesicles were observed to degrade in one of two ways (discussed in detail elsewhere).12 Either the inner bilayer failed, leading to fusion of the two compartments, or the external bilayer failed whereupon the compartment content was released to the external solution.
The above IVTT experiments had to be carried out in different conditions to those commonly used and recommended, specifically in the presence of sucrose and lipids. To test the effect of these additives, we measured GFP expression levels on a fluorimeter with and without these components present. The kit was assembled as before, incubated at 37° for two hours, and fluorescence levels then recorded (Fig. 3). These results revealed that the presence of lipid did not have a detrimental effect on IVTT efficiency. In contrast, the presence of sucrose decreased the reaction efficiency, possibly due to sucrose being a source of RNase contamination.
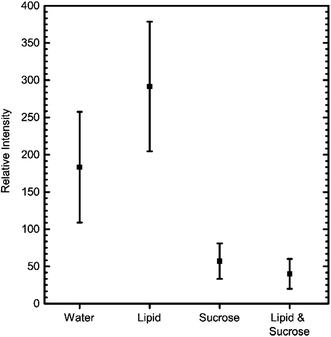 | ||
| Fig. 3 Comparison of the effect different additives have on the efficiency of GFP IVTT, after 2 hours of incubation at 37 °C. Error bars = S.D, n = 5. | ||
In this work, we monitored successful IVTT of different proteins in different compartments via the production of fluorescent proteins. Others have adopted similar approaches to investigate various features of cell-free expression in vesicles, including the stochastic nature of IVTT,17 the effect of vesicle volume on the reconstitution of transmembrane proteins,18 the presence of feedback loops,19 for programmable triggering of IVTT via vesicle fusion,20 for the in vitro evolution of protein pores,21 and for quantitative and statistical insights into proteins synthesis rates.22,23
This work demonstrates the suitability of the multi-compartment vesicle platform to host and effectively compartmentalise distinct, complex biochemical systems such as IVTT, thus further establishing its use as a chassis for vesicle-based artificial cells. This is made possible by the 100% encapsulation efficiency and the ability to finely control the number of compartments and their contents. The method developed thus complements alternative compartmentalisation strategies—including vesicles-in-vesicles,24,25 phase separated vesicles,26,27 and tether-linked vesicles28,29—that have previously proved powerful in a microreactor context.
The fact that our vesicles are larger than biological cells render them unsuitable for certain applications in their current form. However, there are intense ongoing efforts surrounding microfluidic generation of vesicles and related membrane-based structures that enable scale-down in size and a scale-up in generation throughput, which could in principle help facilitate a shift towards functional use in vivo.30
Finally, in this work we extend the concept of soft-matter compartmentalisation to the field of artificial cells.31 Compartmentalisation is a recurring feature in chemical, biological, and engineering systems. It refers to the splitting-up of a unit into distinct spatial sub-divisions, each one considered a module designed separately and optimised to perform a designated task, often in different chemical environments. This enables multi-part, higher-order, or emergent features to arise, and leads to more efficient systems. Our approach differs from bulk self-assembly approaches31 with respect to the greater degree of control over vesicle properties (compartment content, number, and size) and that only one bilayer separates compartments from one another. There is therefore scope for engineering communication routes between compartments, for example by the insertion of transmembrane proteins or DNA origami pores.32
Conclusions
We generated two-compartment vesicles, with two ‘genomes’ contained in each compartment. Using compartmentalisation as an engineering principle allowed protein synthesis of distinct proteins to occur in defined regions, and demonstrated the spatial segregation of complex processes in a vesicle-based artificial cell. This has implications in microreactor and medicinal settings, for instance for isolated on-board synthesis multiple therapeutic biomolecules in a cell-like drug-delivery vehicle, and can be considered an enabling technology for bottom up synthetic biology. Finally, the developed system extends the concept of modularisation that is central to synthetic biology.33,34 Instead of applying to genetic circuits, here it pertains to modular processes that take place within individual compartments of an artificial cell. This paves the way higher-order cellular characteristics to be introduced in future.This work was supported by the EPSRC via grants EP/J017566/1, EP/K038648/1, EP/G00465X/1, BBSRC Grant BB/F013167/1, by an EPSRC Centre for Doctoral Training Studentship from the Institute of Chemical Biology (Imperial College London) awarded to Y.E, and by an EPSRC Doctoral Prize Fellowship awarded to Y.E.
Notes and references
- K. Kurihara, M. Tamura, K.-i. Shohda, T. Toyota, K. Suzuki and T. Sugawara, Nat. Chem., 2011, 3, 775–781, DOI:10.1038/nchem.1127.
- V. Noireaux and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17669–17674 CrossRef CAS PubMed.
- P. Stano, G. Rampioni, P. Carrara, L. Damiano, L. Leoni and P. L. Luisi, BioSystems, 2012, 109, 24–34 CrossRef CAS PubMed.
- P. M. Gardner, K. Winzer and B. G. Davis, Nat. Chem., 2009, 1, 377–383 CrossRef CAS PubMed.
- D. Miller, P. J. Booth, J. M. Seddon, R. H. Templer, R. V. Law, R. Woscholski, O. Ces and L. M. Barter, J. R. Soc., Interface, 2013, 10, 20130496 CrossRef PubMed.
- Y. Elani, A. deMello, X. Niu and O. Ces, Lab Chip, 2012, 12, 3514–3520 RSC.
- P. Carrara, P. Stano and P. L. Luisi, ChemBioChem, 2012, 13, 1497–1502 CrossRef CAS PubMed.
- M. Hadorn, E. Boenzli, K. T. Sørensen, D. De Lucrezia, M. M. Hanczyc and T. Yomo, Langmuir, 2013, 29, 15309–15319 CrossRef CAS PubMed.
- Y. Elani, S. Purushothaman, P. J. Booth, J. Seddon, N. J. Brooks, R. V. Law and O. Ces, Chem. Commun., 2015 10.1039/C5CC00712G.
- P. Janmey and P. Kinnunen, Trends Cell Biol., 2006, 16, 538–546 CrossRef CAS PubMed.
- P. Walde, K. Cosentino, H. Engel and P. Stano, ChemBioChem, 2010, 11, 848–865 CrossRef CAS PubMed.
- Y. Elani, A. Gee, R. V. Law and O. Ces, Chem. Sci., 2013, 4, 3332–3338 RSC.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305, DOI:10.1038/ncomms6305.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS PubMed.
- M. S. Friddin, H. Morgan and M. R. de Planque, Biomicrofluidics, 2013, 7, 014108 CrossRef PubMed.
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. L. Hwang, R. Syeda, J. Thompson and M. Wallace, Mol. BioSyst., 2008, 4, 1191–1208 RSC.
- K. Nishimura, S. Tsuru, H. Suzuki and T. Yomo, ACS Synth. Biol., 2014 DOI:10.1021/sb500249g.
- H. Soga, S. Fujii, T. Yomo, Y. Kato, H. Watanabe and T. Matsuura, ACS Synth. Biol., 2013, 3, 372–379 CrossRef PubMed.
- S. Kobori, N. Ichihashi, Y. Kazuta and T. Yomo, Mol. BioSyst., 2013, 9, 1282–1285 RSC.
- F. Caschera, T. Sunami, T. Matsuura, H. Suzuki, M. M. Hanczyc and T. Yomo, Langmuir, 2011, 27, 13082–13090 CrossRef CAS PubMed.
- S. Fujii, T. Matsuura, T. Sunami, Y. Kazuta and T. Yomo, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 16796–16801 CrossRef CAS PubMed.
- P. van Nies, Z. Nourian, M. Kok, R. van Wijk, J. Moeskops, I. Westerlaken, J. M. Poolman, R. Eelkema, J. H. van Esch and Y. Kuruma, ChemBioChem, 2013, 14, 1963–1966 CrossRef CAS PubMed.
- Z. Nourian, W. Roelofsen and C. Danelon, Angew. Chem., 2012, 124, 3168–3172 CrossRef PubMed.
- H.-P. M. de Hoog, M. Nallani and N. Tomczak, Soft Matter, 2012, 8, 4552–4561 RSC.
- P. Y. Bolinger, D. Stamou and H. Vogel, Angew. Chem., Int. Ed., 2008, 47, 5544–5549 CrossRef CAS PubMed.
- R. Dimova and R. Lipowsky, Soft Matter, 2012, 8, 6409–6415 RSC.
- M. S. Long, A.-S. Cans and C. D. Keating, J. Am. Chem. Soc., 2008, 130, 756–762 CrossRef CAS PubMed.
- A. Jesorka, N. Stepanyants, H. Zhang, B. Ortmen, B. Hakonen and O. Orwar, Nat. Protoc., 2011, 6, 791–805 CrossRef CAS PubMed.
- A. Karlsson, K. Sott, M. Markström, M. Davidson, Z. Konkoli and O. Orwar, J. Phys. Chem. B, 2005, 109, 1609–1617 CrossRef CAS PubMed.
- D. van Swaay, Lab Chip, 2013, 13, 752–767 RSC.
- P. A. Beales and T. K. Vanderlick, Adv. Colloid Interface Sci., 2014, 207, 290–305 CrossRef CAS PubMed.
- N. A. Bell and U. F. Keyser, FEBS Lett., 2014, 588, 3564–3570 CrossRef CAS PubMed.
- P. E. Purnick and R. Weiss, Nat. Rev. Mol. Cell Biol., 2009, 10, 410–422 CrossRef CAS PubMed.
- E. Andrianantoandro, S. Basu, D. K. Karig and R. Weiss, Mol. Syst. Biol., 2006, 2, 2006.0028, DOI:10.1038/msb4100073.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods. See DOI: 10.1039/c4cp05933f |
| This journal is © the Owner Societies 2015 |