Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembly of the anti-fungal polyene amphotericin B into giant helically-twisted nanotapes†
Ian William
Hamley
*a,
Steven
Kirkham
a,
Radoslaw M.
Kowalczyk
a,
Valeria
Castelletto
a,
Mehedi
Reza
b and
Janne
Ruokolainen
b
aSchool of Chemistry, Pharmacy and Food Biosciences, University of Reading, Whiteknights, Reading, Berkshire RG6 6AD, UK. E-mail: i.w.hamley@reading.ac.uk
bDept. of Applied Physics, Aalto University School of Science, P. O. Box 15100, FI-00076, Finland
First published on 20th October 2015
Abstract
The amphiphilic polyene amphotericin B, a powerful treatment for systemic fungal infections, is shown to exhibit a critical aggregation concentration, and to form giant helically-twisted nanostructures via self-assembly in basic aqueous solution.
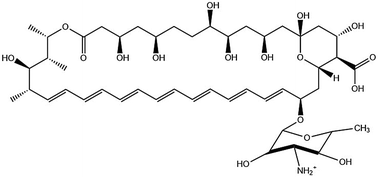
Amphotericin B (AmB, Fig. 1), is an anti-fungal compound in the polyene class that has been used to treat systemic infections for over half a century.1 It was originally isolated in 1955 from a soil culture microbe.1a,c,2 Although it has evaded antimicrobial resistance its toxicity is high, limiting its use at high doses.3 The proposed mode of action involves the formation of pores in lipid membranes facilitated by AmB insertion in the form of dimers.4 The selective antifungal activity results from preferential interaction with ergosterols within fungal cell membranes, compared to cholesterol in mammalian cells.4c,d,5 These dimers result from the facial amphiphilicity of AmB due to pairwise association of the hydrophobic polyene chain (Fig. 1).
The other side of the macrocycle comprises hydrophilic hydroxyl groups, a carboxyl group and a mycosasmine group. It is proposed that ion channels in lipid membranes are maintained in the open form by intermolecular hydrogen bonding between the different hydroxyl, carboxyl and amino groups.
Due to poor solubility in water, AmB is typically formulated with surfactants, and commercial formulations such as Fungizone and AmBisome are widely available.1c,6 Although AmB is poorly soluble in neutral water, it is soluble in acidic and basic aqueous solutions, and in various organic solvents.1a,c,7 The aggregation of AmB under various solution conditions,1d,8 and interacting with lipid membranes4e,8b,c,9 has been studied using spectroscopic methods. Small-angle neutron scattering has also been used to investigate the influence of AmB on lipid membrane structure.10 The influence of different ions across the Hofmeister series on aggregation in aqueous solution has been investigated using UV absorbance and CD measurements.11 Recent spectroscopic (UV fluorescence and circular dichroism, CD) studies point to interesting aggregation properties in pH 12 aqueous solution.1d These studies on aggregation in different environments (aqueous and DMSO solutions) indicate that AmB is present in two different dimer forms, parallel and anti-parallel.1d,8c These can then further self-assemble, shown by time evolution CD spectra, to form tetramers.8c The authors suggest that the dimer form of AmB is the one responsible for the toxicity seen of the drug, which can then associate into tetramers responsible for the transmembrane pore.1d,8c The selectivity of the drug for the sterol ergosterol present in fungi compared to cholesterol in humans has also been shown to be dependent on its aggregation state,1b,11,12 and the therapeutic index is dependent on self-assembly.6b,13 This prior work highlights the considerable interest in AmB and its aggregation behaviour. Despite this, the morphology of the self-assembled structures has not, to our knowledge, been examined as yet. Here we show that in pH 12 aqueous solution, AmB aggregates into giant helically twisted tape structures.
We first established that AmB aggregates via a closed association process, leading to a critical aggregation concentration. This was determined through a fluorescence assay, monitoring the self-fluorescence peak of AmB at 475 nm as a function of concentration (Fig. 2). The discontinuity observed at (0.06 ± 0.01) wt% indicates the critical aggregation concentration (cac). Having established for the first time the cac for AmB, we next examined the nature of self-assembled aggregates observed above this concentration and investigated the structure using fibre X-ray diffraction and SAXS. The kinetics of aggregation were investigated using time-resolved CD experiments.
Fig. 3 shows representative cryo-TEM images obtained after equilibration of a pH 12 aqueous solution of AmB for several days. Remarkable highly extended (over several μm) twisted nanotape structures are evident (additional images are shown in ESI,† Fig. S3). The pitch is relatively uniform, in the range 150–300 nm, however switches in pitch are observed even within single fibres (ESI,† Fig. S4). In some regions of the samples the helical tapes coexist with network-like structures or short straight fibrils (ESI,† Fig. S5).
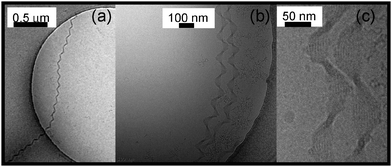 | ||
| Fig. 3 Selection of representative cryo-TEM images showing helically twisted nanotape structures formed by AmB. (a) 1 wt% solution, (b and c) 0.1 wt% solution at pH 11 at different magnifications. | ||
At high magnification (right-hand image in Fig. 3) individual filaments within the nanotapes can be resolved (additional images are shown in ESI,† Fig. S6). Along with the X-ray data to be discussed shortly, this enables a detailed model for the hierarchical self-assembly of AmB molecules to be proposed.
Fibre X-ray diffraction was performed on a stalk dried from a 10 wt% alkaline solution of AmB. The diffraction pattern was found to be isotropic and the circularly integrated intensity profile is shown in Fig. 4.
An approximate unit cell structure was determined by fitting the observed peaks, this indicates an orthorhombic unit cell (P212121) with parameters a = 4.57 Å, b = 10.4 Å, c = 25.3 Å. The c dimension corresponds closely to the estimated molecular length, while b can be assigned to the width of a dimer. Dimer formation of AmB is favoured by facial association of the hydrophobic polyene chains.4c,d Finally, the dimension a is associated with the spacing of the (predominantly planar) molecules. We highlight that this XRD pattern was measured under higher concentration conditions than the cryo-TEM studies, since it was not possible to make stalks from lower concentration solutions which were too fluid. However, it was possible to perform SAXS in solution under concentration conditions 0.5–2 wt%, covering the range of the conditions of the cryo-TEM images. The corresponding profiles are shown in ESI,† Fig. S7. These profiles show a Bragg peak corresponding to d = 17.9 Å which differs from the spacings in Fig. 4, perhaps due to changes in nanostructure with concentration (also the pH was slightly lower for the SAXS measurements). The dependence of nanostructure on pH and concentration is currently under further investigation.
Consistent with the cryo-TEM and XRD studies we propose the model shown in Fig. 5 for the aggregation of AmB at pH 12. The energy minimized (MM2 in Chem3D Pro) conformation of the molecule is a twisted “chair-like” conformation (Fig. 5) which leads to twisting of the filaments seen within the nanotapes. The molecules probably aggregate via pairwise association of hydrophobic and hydrophilic faces, as discussed above. The exact nature of the dimeric species (i.e. parallel vs. antiparallel) is not currently known, so dimers are not illustrated in Fig. 5. What can be deduced from the high magnification cryo-TEM images is that the filament spacing is approximately 5 nm, and the filaments themselves are 2–3 nm wide, which is close to the molecular length as indicated in the tentative proposed model in Fig. 5.
The kinetics of aggregation above the cac were investigated using circular dichroism spectroscopy. AmB produces a CD spectrum rich in features as it is a chiral molecule with a complex UV absorbance spectrum due to the presence of the polyene chain. Fig. 6 shows representative CD spectra recorded for AmB as a function of time after sample preparation for a 0.1 wt% sample, data being measured at 20 °C.
These spectra are consistent with AmB being present in the state of aggregates. Previous reports8a,11,14 assign UV absorbance features in the 320–360 nm range to aggregates, whereas monomers give a pronounced vibronic peak at 409 nm, not seen in the spectra in Fig. 6. The development of the strong negative CD peak at 326 nm, not previously reported, is ascribed to the formation of the giant helical tapes shown in Fig. 3. The rest of the CD spectrum is similar to that previously reported for AmB in dilute aqueous solutions.1d,11,14 Auto-oxidation of AmB due to the production of reactive oxygen species has previously been noted in DMSO/buffer,12 and DMSO/aqueous solutions.14 We therefore obtained mass spectroscopy and 1H and 13C NMR spectra for aged solutions of AmB. These are presented in ESI,† Fig. S1 and S2, respectively and provide evidence that no molecular degradation or auto-oxidation process has occurred. The electrospray mass spectrum shows peaks (charge z = 1 and z = 2) corresponding to the expected molar mass, and the 1H and 13C NMR spectrum can be assigned to the expected structure shown in Fig. 1. NMR assignments were based on previously reported spectra (in DMSO).15
The twisted nanotapes formed by AmB appear to comprise a quite well defined number of strands (Fig. 3c), approximately ten (this is subject to uncertainty due to depth resolution limitations in the cryo-TEM images). The selection of a finite number of filaments and of the helical pitch in twisted peptide ribbon assemblies has been rationalised quantitatively based on models that account for the interplay between electrostatic and either attractive dispersive interactions or mechanical properties (torsional elastic energy).16 Computer simulations of helical filament assembly have also been reported recently.17 In future measurements, we plan to obtain the Young's modulus of AmB fibrils in order to make more quantitative comparison with models which rely on knowledge of elastic properties. The model of Nyrkova and Semenov et al.18 is based on the assembly of stacks of helical ribbons which does not seem applicable to the AmB fibres which appear to comprise arrays of cylindrical filaments (Fig. 5).
In summary, we report the formation of giant helical nanotapes resulting from the self-assembly of amphotericin B in pH 12 aqueous solution. The nanotapes comprise an array of helical filaments. There is an associated strong feature in the UV CD spectrum resulting from the chiral molecular arrangement. We also show that AmB exhibits a critical aggregation concentration in pH 12 aqueous solution and provide a unit cell structure from fibre X-ray diffraction.
This work was supported by EPSRC Platform Grant EP/L020599/1 to IWH. Use of the Chemical Analysis Facility at the University of Reading is gratefully acknowledged. We thank Dr Katsuaki Inoue for assistance during SAXS beamtime at Diamond (ref. SM12321).
Notes and references
- (a) I. M. Asher, G. Schwartzman and USASRG, Anal. Profiles Drug Subst., 1977, 6, 1–42 CrossRef CAS; (b) H. A. Gallis, R. H. Drew and W. W. Pickard, Rev. Infect. Dis., 1990, 12, 308–329 CrossRef CAS PubMed; (c) J. J. Torrado, R. Espada, M. P. Ballesteros and S. Torrado-Santiago, J. Pharm. Sci., 2008, 97, 2405–2425 CrossRef CAS PubMed; (d) J. Starzyk, M. Gruszecki, K. Tutaj, R. Luchowski, R. Szlazak, P. Wasko, W. Grudzinski, J. Czub and W. I. Gruszecki, J. Phys. Chem. B, 2014, 118, 13821–13832 CrossRef CAS PubMed.
- J. Vandeputte, J. L. Wachtel and E. T. Stiller, Antibiot. Annu., 1955, 3, 579–587 Search PubMed.
- S. A. Davis, B. M. Vincent, M. M. Endo, L. Whitesell, K. Marchillo, D. R. Andes, S. Lindquist and M. D. Burke, Nat. Chem. Biol., 2015, 11, 481–487 CrossRef CAS PubMed.
- (a) B. de Kruijf, W. Gerritse, A. Oerlemans, A. R. A. Demel and L. L. M. van Deenen, Biochim. Biophys. Acta, 1974, 339, 30–43 CrossRef; (b) B. de Kruijf and R. A. Demel, Biochim. Biophys. Acta, 1974, 339, 57–70 CrossRef; (c) J. Milhaud, V. Ponsinet, M. Takashi and B. Michels, Biochim. Biophys. Acta, Biomembr., 2002, 1558, 95–108 CrossRef CAS; (d) G. Barratt and S. Bretagne, Int. J. Nanomed., 2007, 2, 301–313 CAS; (e) M. Gagoś and M. Arczewska, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 2124–2130 CrossRef PubMed.
- C. K. Kang, K. Yamada, Y. Usuki, A. Ogita, K. Fujita and T. Tanaka, Microbiology, 2013, 159, 939–947 CrossRef CAS PubMed.
- (a) A. H. A. Mohamed-Ahmed, K. A. Les, K. Seifert, S. L. Croft and S. Brocchini, Mol. Pharmaceutics, 2013, 10, 940–950 CrossRef CAS PubMed; (b) J. A. Sanchez-Brunete, M. A. Dea, S. Rama, F. Bolas, J. M. Alunda, R. Raposo, M. T. Mendez, S. Torrado-Santiago and J. J. Torrado, Antimicrob. Agents Chemother., 2004, 48, 3246–3252 CrossRef CAS PubMed.
- M. J. O'Neil, The Merck Index, Cambridge, UK, 2013 Search PubMed.
- (a) W. I. Gruszecki, M. Gagos and M. Herec, J. Photochem. Photobiol., B, 2003, 69, 49–57 CrossRef CAS; (b) W. I. Gruszecki, R. Luchowski, M. Gagos, M. Arczewska, P. Sarkar, M. Herec, B. Mysliwa-Kurdziel, K. Strzalka, I. Gryczynski and Z. Gryczynski, Biophys. Chem., 2009, 143, 95–101 CrossRef CAS PubMed; (c) P. Wasko, R. Luchowski, K. Tutaj, W. Grudzinski, P. Adamkiewicz and W. I. Gruszecki, Mol. Pharmaceutics, 2012, 9, 1511–1520 CrossRef CAS PubMed.
- J. Mazerski, J. Bolard and E. Borowski, Biochem. Biophys. Res. Commun., 1983, 116, 520–526 CrossRef CAS PubMed.
- (a) M. Herec, A. Islamov, A. Kuklin, M. Gagos and W. I. Gruszecki, Chem. Phys. Lipids, 2007, 147, 78–86 CrossRef CAS PubMed; (b) F. Foglia, A. F. Drake, A. E. Terry, S. E. Rogers, M. J. Lawrence and D. J. Barlow, Biochim. Biophys. Acta, Biomembr., 2011, 1808, 1574–1580 CrossRef CAS PubMed; (c) F. Foglia, S. E. Rogers, J. R. P. Webster, F. A. Akeroyd, K. F. Gascoyne, M. J. Lawrence and D. J. Barlow, Langmuir, 2015, 31, 8042–8051 CrossRef CAS PubMed.
- M. T. Grijalba, M. Cheron, E. Borowski, J. Bolard and S. Schreier, Biochim. Biophys. Acta, Gen. Subj., 2006, 1760, 973–979 CrossRef CAS PubMed.
- M. T. Lamyfreund, V. F. N. Ferreira, A. Faljonialario and S. Schreier, J. Pharm. Sci., 1993, 82, 162–166 CrossRef CAS.
- (a) C. Petit, M. Cheron, V. Joly, J. M. Rodrigues, J. Bolard and F. Gaboriau, J. Antimicrob. Chemother., 1998, 42, 779–785 CrossRef CAS PubMed; (b) B. Cybulska, I. Gadomska, J. Mazerski, J. Grzybowska, E. Borowski, M. Cheron and J. Bolard, Acta Biochim. Pol., 2000, 47, 121–131 CAS.
- F. Gaboriau, M. Chéron, L. Leroy and J. Bolard, Biophys. Chem., 1997, 66, 1–12 CrossRef CAS PubMed.
- (a) C. M. McNamara, S. Box, J. M. Crawforth, B. S. Hickman, T. J. Norwood and B. J. Rawlings, J. Chem. Soc., Perkin Trans. 1, 1998, 83–87 RSC; (b) B. Murphy, K. Anderson, C. Borissow, P. Caffrey, G. Griffith, J. Hearn, O. Ibrahim, N. Khan, N. Lamburn, M. Lee, K. Pugh and B. Rawlings, Org. Biomol. Chem., 2010, 8, 3758–3770 RSC.
- (a) J. Adamcik and R. Mezzenga, Soft Matter, 2011, 7, 5437–5443 RSC; (b) I. Usov and R. Mezzenga, ACS Nano, 2014, 8, 11035–11041 CrossRef CAS PubMed; (c) S. Assenza, J. Adamcik, R. Mezzenga and P. De Los Rios, Phys. Rev. Lett., 2014, 113 Search PubMed.
- M. Gruziel and P. Szymczak, Soft Matter, 2015, 11, 6294–6304 RSC.
- (a) I. A. Nyrkova, A. N. Semenov, A. Aggeli and N. Boden, Eur. Phys. J. B, 2000, 17, 481–497 CrossRef CAS; (b) I. A. Nyrkova, A. N. Semenov, A. Aggeli, M. Bell, N. Boden and T. C. B. McLeish, Eur. Phys. J. B, 2000, 17, 499–513 CrossRef CAS; (c) A. Aggeli, I. A. Nyrkova, M. Bell, R. Harding, L. Carrick, T. C. B. McLeish, A. N. Semenov and N. Boden, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 11857–11862 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Methods, mass spectroscopy and NMR data, cryo-TEM images and SAXS data. See DOI: 10.1039/c5cc08224b |
| This journal is © The Royal Society of Chemistry 2015 |