Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assessing the effect of different shapes of glyco-gold nanoparticles on bacterial adhesion and infections†
Preeti Madhukar
Chaudhary‡
a,
Sivakoti
Sangabathuni‡
a,
Raghavendra Vasudeva
Murthy
a,
Ajay
Paul
b,
Hirekodathakallu V.
Thulasiram
b and
Raghavendra
Kikkeri
*a
aIndian Institute of Science Education and Research, Pashan, Pune 411008, India. E-mail: rkikkeri@iiserpune.ac.in; Fax: +91-20-25899790; Tel: +91-20-25908207
bOrganic Chemistry, CSIR-National Chemical Laboratory, Dr. Homi Bhabha Road, Pune 411008, India
First published on 28th August 2015
Abstract
Achieving selective and sensitive carbohydrate–protein interactions (CPIs) using nanotechnology is an intriguing area of research. Here we demonstrate that the different shapes of gold nanoparticles (AuNPs) functionalized with monosaccharides tune the bacterial aggregations. The mechanism of aggregation revealed that the large number of surface interactions of rod shaped mannose-AuNPs with E. coli ORN 178 compared with spherical and star-shaped AuNPs exhibited higher avidity and sensitivity. Moreover, such sensitive binding can be used for effective inhibition of bacterial infection of cells.
Carbohydrate–protein interactions (CPIs) are one of the most important and major events on cell surfaces.1 The basic components involved in the interaction are the cell surface glycans, which demonstrate a sensitive and selective cis/trans binding with protein counterparts. However, due to weak CPIs, nature facilitates multivalency in target specific proteins. Recently, extensive efforts have been directed toward mimicking these bio-events by replicating the multivalent scaffolds.2 However, to fully understand CPIs, it is important to quantify binding affinity with different sizes, shapes, orientations, and local concentrations of the sugars.3 Varieties of synthetic multivalent templates such as cyclodextrins,4 calixarenes,5 polymers,6 dendrimers,7 and supramolecular complexes8 have been extensively used to decorate sugars in particular topology to target CPIs. These synthetic templates offer controllability and reproducibility, but the drawbacks include production methods that sometimes require complex synthetic conditions. Another class of templates for multivalent glyco-probes are nanoparticles (NPs), such as gold, silver, iron and CdSe NPs.9 These templates are more rigid and easy to synthesize in large quantities in different sizes, shapes and orientations and can also be easily decorated chemically and biologically.11
Among these NPs, AuNPs are more attractive for their different shapes (rod, sphere, star, cube and spindle), surface resonance band and ease of characterization by UV-visible and transmission electron microscopy (TEM).10 Moreover, they are less toxic compared to quantum dots. Penadés and co-workers used AuNPs to decorate more than one sugar to target HIV, bacteria, modulation of immune responses, and CPIs.11 Similarly, enormous effort has been expended by the group of Lin, Wu, Tseng and others to develop gold nanodots and nanoclusters for the sensitive and selective detection of E. coli.12 However, in all these studies, the shapes of AuNPs were kept constant to validate the binding with bacteria, cells or organs, limiting the assessment of the role of the different shapes of nanoparticles involved in specific CPIs. The latter is of fundamental importance for understanding the CPIs and developing new biomaterials. Various targeting units, such as antibodies, peptides, aptamers have been functionalized on different shapes of AuNPs to enhance their specificity for tumors, immune responses and biosensing processes.13 However, a systematic investigation of shape dependent CPIs with the same volume and sugar density and its potential applications have not been reported.
Herein, we report the use of glyco-AuNps in bacterial recognition and inhibiting bacterial infection. Three different shapes (rod, sphere and star) of gold nanoparticles coated with mannose and galactose sugar substrates and PEG were used to quantify the binding affinity with E. coli. To profile the potential applications of the shape dependent CPIs, the inhibition of E. coli infection of HeLa cells was quantified.
The monovalent mannose- and galactose-modified linkers (7 and 8) were synthesized by using the slightly modified procedure from the protocol reported in the literature.14 Briefly, the conjugation of triethylene glycol with 11-bromoundec-1-ene, followed by glycosylation with per-acetylated mannose or galactose yielded compound 3 or 4. These compounds were further treated with thioacetic acid in the presence of azo-isobutyronitrile (AIBN), followed by deacetylation with NaOMe to yield compound 7 or 8. (Scheme 1).
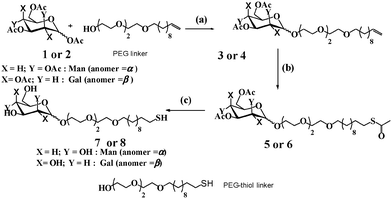 | ||
| Scheme 1 Synthesis of mannose and galactose modified linker: (a) BF3·Et2O/DCM; (b) thioaceticacid/AIBN/dioxane/60 °C; (c) NaOMe/MeOH. | ||
Rod AuNPs coated with cetyltrimethylammonium bromide (CTAB) were synthesized via a seeding growth method as reported previously (Scheme 2).15 The synthesis of star shaped AuNPs was carried out using the biocompatible surfactant free method16 and spherical AuNPs by reduction of chloroauric acid with sodium citrate.17 Finally, sugar encapsulation of AuNPs was carried out by a ligand exchange process.
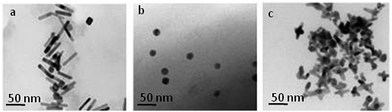
The physical characteristics of AuNP complexes are presented in Table 1. The shape and size of the synthesized AuNPs were confirmed by scanning electron microscopy (SEM) (Fig. S2, ESI†), transmission electron microscopy (TEM) (Fig. 1 and Fig. S3, ESI†) and UV-vis absorption (Fig. S1, ESI†). Spherical AuNPs (18.5 ± 2.5), nano-rods (45.6 ± 3.5 × 11.2 ± 0.5) and nano-stars (40.6 ± 3.1 × 17 ± 0.9) of equal volume were used as model particles in this study. AuNPs functionalized with sugar and PEG was confirmed by changes in the zeta (ζ) potential (Table 1). After sugar ligation, the ζ-potentials for rod AuNPs changed from positive to negative (34 to −10 mV), indicating the effective ligand changes of CTAB surfactant by the sugar substrates. At the same time, spherical and star shaped AuNPs showed only a slight change in the negative potential (sphere: −24 to −17 mV; star: −29 to −23 mV). This may be due to the displacement of negatively charged citrate and HEPES surfactant by sugar scaffolds respectively.
| Particle | Diameter(s) (nm) | λ max (nm) | ζ-potentials (mV) |
|---|---|---|---|
| Rod | 45.6 ± 3.5 × 11.3 ± 0.5 | 798 | 34.4 ± 0.3 |
| Sphere | 18.5 ± 2.5 | 525 | −24 ± 0.2 |
| Star | 40.6 ± 3.1 × 17.1 ± 0.9 | 715 | −29 ± 1.4 |
| Man-rod | 45.9 ± 3.2 × 11.5 ± 1.7 | 799 | −10.7 ± 0.5 |
| Gal-rod | 45.2 ± 2.2 × 11.8 ± 0.7 | 802 | −17.3 ± 0.8 |
| PEG-rod | 45.9 ± 3.1 × 11.3 ± 1.5 | 798 | −30.1 ± 0.6 |
| Man-sphere | 20.5 ± 1.3 | 520 | −21.3 ± 0.5 |
| Gal-sphere | 20.5 ± 1.2 | 525 | −17.5 ± 0.3 |
| PEG-sphere | 19.2 ± 1.8 | 525 | −15.3 ± 0.2 |
| Mann-star | 44.2 ± 0.2 × 18.2 ± 0.9 | 725 | −22.4 ± 1.1 |
| Gal-star | 42.9 ± 0.2 × 18.2 ± 0.7 | 725 | −23.8 ± 1.3 |
| PEG-star | 44.6 ± 0.2 × 17.4 ± 0.9 | 725 | −16.2 ± 1.6 |
The number of sugar molecules on AuNPs was quantified by using a phenol–sulfuric acid method (Table S1, ESI†). As expected, almost equal concentration of sugar scaffolds were confirmed. Experiments were performed using two E. coli strains to validate the shape dependent bacterial adhesion. The choice of bacterial strains was based on their ability to recognize specific sugar substrates. In this study, we choose FimH–mannose interactions based on ORN 178 and ORN 208 is a mutant having no FimH receptors.4a Fluorescence imaging clearly showed aggregation of bacteria due to specific CPIs.
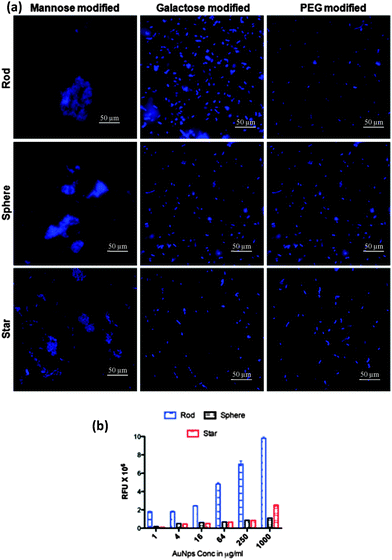
Our first experiment was to establish the binding selectivity. In this experiment, we incubated different shapes and sugar conjugated AuNPs with ORN 178 and ORN 208 for 1 h in PBS solution. After that cells were centrifuged and the aggregates formed by the bacteria were imaged and quantified. As expected, the maximum number of aggregation was observed with mannose coated AuNPs compared to the galactose and PEG counterparts (Fig. 2a). On the contrary, no aggregation formation was seen in ORN 208 in any of AuNPs (Fig. S4, ESI†). Thus, the difference in specific vs. nonspecific binding was highly dependent on the sugar scaffold compared to the shape of the AuNPs. Although all three different shapes of AuNPs contained the same amount of sugar, they apparently differ in three important physical properties such as: (1) contact area of the NPs with respect to external stimuli; (2) rotational volume availability and; and (3) aspect ratio. Thus, we hypothesize that a specific shape might induce sensitive bacterial adhesion.
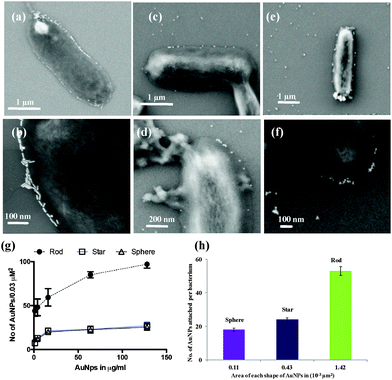
To demonstrate the shape dependent bacterial adhesion, experiments were performed using different concentrations of rod, sphere and star shaped Man-AuNPs (Fig. S5, ESI†). The dose–response bar graph of different shapes is presented in Fig. 2b. Two groups of interactions could be identified in terms of their aggregation. The first group, composed of spherical and star shaped AuNPs, gave a detection limit of 17 ± 2 μg ml−1 and 14 ± 2 μg ml−1 respectively with a known number of ORN 178 E. coli strain. The second group, composed of rod-shaped AuNPs, gave a detection limit of 0.03 ± 0.01 μg ml−1 under the same conditions. The observed ∼80-fold difference in the detection of bacteria by rod AuNPs could be attributed to several factors: (1) aspect ratio, which could increase specific attachment of particles on bacterial cell surfaces; (2) the effective surface area available for AuNPs binding; and (3) self-assembly of NPs to amplify the specific interaction. To further validate the above results, SEM images were obtained at the lowest concentration (0.8 μg ml−1) of AuNP treated bacteria (Fig. S7–S9, ESI†). As expected, the relative amount of mannose rod AuNPs involved in effective CPIs is higher than spherical and star shaped counterparts. This might be because of the interaction of the large number of rod AuNPs with the E. coli surface compared to star shaped or spherical counterparts (Fig. 3a and b).18 Upon close examination of the rod shaped AuNPs, we observed head-to-head self-assembly on the surface of the bacteria (Fig. S7, ESI†). In contrast, star and spherical AuNPs under the same conditions resulted in mono-dispersed and very few aggregations, indicating that the aspect ratio of AuNPs is crucial for self-assembly and to improve the sensitive interactions (Fig. 3c–f) we also quantified the number of nanoparticles adhered on a specific μm2 area of the bacteria (Fig. 3g). The binding density of rod AuNPs was found to be nearly 2.5-fold higher than that of star and spherical shaped AuNPs. This confirmed that the rod AuNPs occupied more surface area on bacteria than that of star and spherical AuNPs, which directly modulate the binding affinity. All these results are correlated with the mathematical model and flow chemistry experiment proposed by Kohlar et al.19 Finally, we quantified the number of AuNPs actively bound on the surface of the bacteria. We observed that spherical AuNPs occupied nearly 10- and 4-fold less surface area of the bacteria compare to rod and star shaped AuNPs, indicating that rod AuNPs exhibit higher avidity toward the bacteria (Fig. 3h).
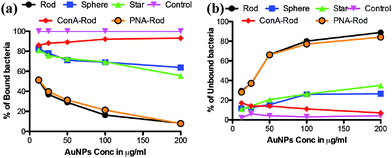
To demonstrate the potential for application of shape dependent CPIs, HeLa cells, which have been known to express high mannose on the cell surface20 were infected with a known amount of DAPI stained ORN 178 E. coli. We examined infection caused by the bacteria in the presence of three different shapes and at five different concentrations (12, 25, 50, 100 and 200 μg ml−1) of mannose-AuNPs. After 1 h of incubation, both bound and unbound bacteria were separated and quantified by measuring the fluorescence intensity of DAPI. The results obtained from different shapes showed that the percentage of inhibition (unbound bacteria) caused by rod AuNPs was approximately six-fold more than that of the star and spherical AuNPs (Fig. 4). To confirm that the inhibition effect of mannose-rod AuNPs is indeed anchored by specific CPIs, ConA and PNA (10 μl of 0.1 mg ml−1) were added to mannose-AuNPs before mixing with bacteria and HeLa cells. Due to specific CPIs between mannose-ConA most of the cells got infected, whereas in the presence of PNA lectin, which is specific to galactose sugars, effective inhibition of cell–bacterial interactions was seen. These results confirmed that the shape of the AuNPs and specific CPIs fine-tune the inhibition properties.
In summary, we have demonstrated that shape is one of the factors in establishing sensitive carbohydrate–protein interactions. Data from the bacterial adhesion and HeLa cell infection studies showed that the rod-shaped AuNPs functionalized with mannose had substantial sensitivity compared to that of star-shaped and spherical shaped AuNPs. Factors such as self-assembly and effective surface contact are critical for sensitive adhesion. In a more general perspective, blockage of E. coli infection by rod mannose-AuNPs may open opportunities for the development of efficient medicines for urinary or digestive tract infections.4,9c
R.K, and P.C. thank CSIR-RA, IISER Pune, Indo-German (DST-MPG) program and DST (Grant No. SB/S1/C-46/2014) and UGC, India, for financial support. The mutants E. coli ORN 178 and ORN 208 were provided by Prof. Orndorff (College of Veterinary Medicine, Raleigh, NC United States).
References
- (a) M. Cohen and A. Varki, Int. Rev. Cell Mol. Biol., 2014, 308, 75 CrossRef CAS PubMed; (b) C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357 CrossRef CAS; (c) H. Lis and N. Sharon, Chem. Rev., 1998, 98, 637 CrossRef CAS PubMed.
- (a) Y. M. Chabre and R. Roy, Chem. Soc. Rev., 2013, 42, 4657 RSC; (b) M. Gingras, Y. M. Chabre, M. Roy and R. Roy, Chem. Soc. Rev., 2013, 42, 4823 RSC; (c) P. Bojarová, R. R. Rosencrantz, L. Elling and V. Křen, Chem. Soc. Rev., 2013, 42, 4774 RSC.
- R. J. Pieters, Org. Biomol. Chem., 2009, 7, 2013 CAS.
- (a) M. Gade, A. Paul, C. Alex, D. Choudhury, H. V. Thulasiram and R. Kikkeri, Chem. Commun., 2015, 51, 6346 RSC; (b) S. venwetswinkel, A. N. Volkov, Y. G. Sterckx, A. Garcia-Pino, L. Buts, W. F. Vranken, J. Bouckaert, R. Roy, L. Wyns and N. A. van Nuland, J. Med. Chem., 2014, 57, 1416 CrossRef PubMed.
- (a) L. Baldini, A. Casnati, F. Sansone and R. Ungaro, Chem. Soc. Rev., 2007, 36, 254 RSC; (b) J. D. Badjić, A. Nelson, S. J. Cantrill, W. B. Turnbull and J. F. Stoddart, Acc. Chem. Res., 2005, 38, 723 CrossRef PubMed.
- (a) L. L. Kiessling and J. C. Grim, Chem. Soc. Rev., 2013, 42, 4476 RSC; (b) L. R. Prost, J. C. Grim, M. Tonelli and L. L. Kiessling, ACS Chem. Biol., 2012, 7, 1603 CrossRef CAS PubMed.
- (a) R. Kikkeri, D. Grünstein and P. H. Seeberger, J. Am. Chem. Soc., 2010, 132, 10230 CrossRef CAS PubMed; (b) R. Yadav and R. Kikkeri, Chem. Commun., 2012, 48, 1704 RSC.
- (a) Y. Kim, S. Shin, T. Kim, D. Lee, C. Seok and M. Lee, Angew. Chem., Int. Ed. Engl., 2013, 52, 6426 CrossRef CAS PubMed; (b) D. Grünstein, M. Maglinao, R. Kikkeri, M. Collot, K. Barylyuk, B. Lepenies, F. Kamena, R. Zenobi and P. H. Seeberger, J. Am. Chem. Soc., 2011, 133, 13957 CrossRef PubMed.
- (a) N. C. Reichardt, M. Martín-Lomas and S. Penadés, Chem. Soc. Rev., 2013, 42, 4358 RSC; (b) M. Marradi, F. Chiodo, I. García and S. Penadés, Chem. Soc. Rev., 2013, 7, 4728 RSC; (c) K. Turcheniuk, C.-H. Hang, J. Spadavecchia, A. Y. Serrano, I. Larroulet, A. Pesquera, A. Zurutuza, M. G. Pisfil, L. Héliot, J. Boukaert, R. Boukherroub and S. Szunerits, J. Mater. Chem. B, 2015, 3, 375 RSC.
- (a) S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209 RSC; (b) S. H. Radwan and H. M. Azzazy, Expert Rev. Mol. Diagn., 2009, 9, 511 CrossRef CAS PubMed.
- (a) F. Chiodo, M. Marradi, J. Park, A. F. J. Ram, S. Penadés, I. van Die and B. Tefsen, ACS Chem. Biol., 2014, 9, 383 CrossRef CAS PubMed; (b) D. Arosio, F. Chiodo, J. J. Reina, M. Marelli, S. Penadés, Y. van Kooyk, J. J. Garcia-Vallejo and A. Bernardi, Bioconjugate Chem., 2014, 25, 2244 CrossRef CAS PubMed; (c) I. García, A. Sanchez-Iglesias, M. Henriksen-Lacey, M. Grzelczak, S. Penades and L. M. Liz-Marzan, J. Am. Chem. Soc., 2015, 137, 3686 CrossRef PubMed; (d) F. Chiodo, M. Marradi, B. Tefsen, H. Snippe, I. van Die and S. Penadés, PLoS One, 2013, 8, e73027 CAS.
- (a) C.-C. Lin, Y.-C. Yeh, C.-Y. Yang, C.-L. Chen, G.-F. Chen, C.-C. Chen and Y.-C. Wu, J. Am. Chem. Soc., 2002, 124, 3508 CrossRef CAS PubMed; (b) Y.-T. Tseng, H.-T. Chang, C.-T. Chen, C.-H. Chen and C.-C. Huang, Biosens. Bioelectron., 2011, 27, 95 CrossRef CAS PubMed; (c) P.-H. Chan, B. Ghosh, H.-Z. Lai, H.-L. Peng, K. K. T. Mong and Y.-C. chen, PLoS One, 2013, 8, e58064 CAS; (d) C.-C. Huang, C.-T. Chen, Y.-C. Shiang, Z.-H. Lin and H.-T. Chang, Anal. Chem., 2009, 81, 875 CrossRef CAS PubMed.
- (a) J. Wang, G. Zhu, M. You, E. Song, M. I. Shukoor, K. Zhang, M. B. altman, Y. Chen, Z. Zhu, C. Z. Huang and W. Tan, ACS Nano, 2012, 6, 5070–5077 CrossRef CAS PubMed; (b) X. yang, X. Liu, Z. Liu, F. Pu, J. Ren and X. Qu, Adv. Mater., 2012, 24, 2890–2895 CrossRef CAS PubMed; (c) L. A. Dykman and N. G. Khlebtson, Chem. Rev., 2014, 114, 1258–1288 CrossRef CAS PubMed; (d) K. Niikura, T. Matsunga, T. Suzuki, S. Kobayashi, H. Yamaguchi, Y. Orba, A. Kawaguchi, H. Hasegawa, K. Kajino, T. Ninomiya, K. Ijiro and H. Sawa, ACS Nano, 2013, 7, 3926 CrossRef CAS PubMed.
- G. Bellapadrona, A. B. Tesler, D. Grünstein, L. H. Hossain, R. Kikkeri, P. H. Seeberger, A. Vaskevich and I. Rubinstein, Anal. Chem., 2012, 84, 232 CrossRef CAS PubMed.
- B. Nikoobakht and M. A. El-Sayed, Chem. Mater., 2003, 15, 1957 CrossRef CAS.
- J. Xie, J. Y. Lee and D. I. C. Wang, Chem. Mater., 2007, 19, 2823 CrossRef CAS.
- E. C. Cho, Y. Liu and Y. Xia, Angew. Chem., Int. Ed., 2010, 49, 1976 CrossRef CAS PubMed.
- W. Li, P. Zhang, M. Dai, J. He, T. Babu, Y.-L. Xu, R. Deng, R. Liang, M.-H. Lu, Z. Nie and J. Zhu, Macromolecules, 2013, 46, 2241 CrossRef CAS.
- P. Kolhar, A. C. Anselmo, V. Gupta, K. Pant, B. Prabhakarpandian, E. Ruoslahti and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 10753 CrossRef CAS PubMed.
- P. M. Chaudhary, R. V. Murthy, R. Yadav and R. Kikkeri, Chem. Commun., 2015, 51, 8112 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cc05238f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |