Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A systematic approach to diverse, lead-like scaffolds from α,α-disubstituted amino acids†
Daniel J.
Foley
a,
Richard G.
Doveston
a,
Ian
Churcher
b,
Adam
Nelson
*ac and
Stephen P.
Marsden
*a
aSchool of Chemistry, University of Leeds, Leeds, LS2 9JT, UK. E-mail: s.p.marsden@leeds.ac.uk; a.s.nelson@leeds.ac.uk
bGlaxoSmithKline Medicines Research Centre, Stevenage, SG1 2NY, UK
cAstbury Centre for Structural Molecular Biology, University of Leeds, Leeds, LS2 9JT, UK
First published on 11th May 2015
Abstract
A powerful strategy for the efficient lead-oriented synthesis of novel molecular scaffolds is demonstrated. Twenty two scaffolds were prepared from just four α-amino acid-derived building blocks and a toolkit of six connective reactions. Importantly, each individual scaffold has the ability to specifically target lead-like chemical space.
There is an intrinsic link between the molecular properties of initial screening compounds and the likelihood of their successful progression towards drug candidates.1,2 Since both molecular weight and lipophilicity tend to increase during the lead optimisation phase,3 controlling the molecular properties of lead molecules is crucial if the final compounds are to remain in drug-like space. However, a recent survey concluded that just 2.6% of 4.9 M commercially-available molecules were lead-like,4 even before compound diversity5 was considered. It has also been shown that compounds with higher fractions of sp3-hybridised atoms have lower attrition rates than less saturated compounds,2 and thus may serve as better leads. Obtaining large numbers of diverse, highly three-dimensional, lead-like small molecules is thus a major challenge in maintaining high-quality screening collections. The concept of lead-oriented synthesis has been proposed4 to frame the challenge of delivering diverse molecules with specific molecular properties of direct relevance to the drug discovery and development process.4,6,7 Lead-oriented syntheses should, inter alia,4 focus on the diversity of structures generated, synthetic efficiency, tolerance to polar functionality, and not be susceptible to ‘Log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P drift’, the phenomenon whereby higher failure rates of more polar compounds in a given array synthesis leads to an increase in mean and median log
P drift’, the phenomenon whereby higher failure rates of more polar compounds in a given array synthesis leads to an increase in mean and median log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for final libraries versus that designed. The significant academic challenge laid down by such demands has recently begun to prompt the syntheses of specific classes of lead-like molecules.8
P for final libraries versus that designed. The significant academic challenge laid down by such demands has recently begun to prompt the syntheses of specific classes of lead-like molecules.8
Making diverse compound libraries is not necessarily difficult per se, but without careful planning comes at the expense of synthetic efficiency and, until recently,8a has lacked a lead-oriented focus. In order to realise efficient lead-oriented synthesis, our approach is to exploit small building blocks that contain multiple chemically-orthogonal functional groups; systematic exploitation of a minimal toolkit of reactions for the pairwise coupling of the functional groups present would rapidly deliver diverse scaffolds that, on decoration of the newly formed or residual functional groups, should target broad regions of lead-like chemical space.
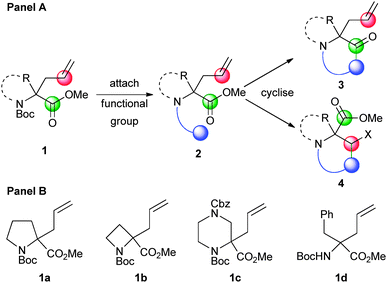
To validate our strategy, we chose readily available9 α-allyl α-amino acid derivatives 1 as the substrates, the amine functionality of which would then be armed with a functional group (blue) to yield a trifunctional intermediate 2, tuning the precursor for cyclisation with either the adjacent ester (green; → 3) or alkene (red; → 4) (Scheme 1). Variation of the resulting scaffold should be possible by exploiting acyclic or cyclic amino acid derivatives (e.g.1a–d) as substrates; by varying the appended functionality (blue); and by varying the cyclisation reaction.
Firstly, allylation of commercial N-Boc-protected amino acid esters furnished the trifunctional building blocks 1.9 Additional intermediates bearing alternative N-substituents were prepared to tune the precursors 1 for cyclisation (Scheme 2, Panel A). Removal of the Boc-protecting group was followed by formation of acyl ureas 6 (treatment with ethyl isocyanatoformate) or by alkylation to give protected 1,2-diamines 9, o-bromobenzylamines 12 and allylamines 15. We then used these substrates to synthesise cyclic molecular scaffolds, by exploiting the use of a toolkit of three broad reaction types, all targeting medicinally relevant motifs.
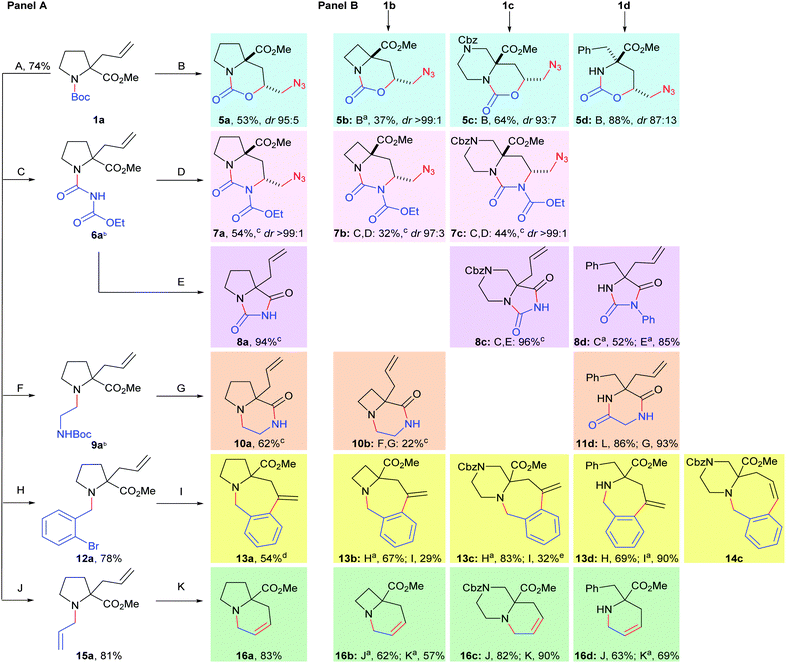 | ||
Scheme 2 Synthesis of diverse lead-like molecular scaffolds. Coloured boxes correspond to different cyclisation reactions, new bonds/groups are shown in red. Panel A: synthesis of scaffolds from 1a. Panel B: extension to alternative scaffolds using building blocks 1b–d. Standard methods: A: TFA, CH2Cl2; B: (i) I2, THF–H2O; (ii) NaN3, DMF; C: (EtO2C)NCO, CH2Cl2; D: (i) I2, Li[Al(OtBu)4], THF, −5 °C; (ii) NaN3, DMF; E: NaOMe, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 MeOH–PhMe, 65 °C; F: N-Boc-glycinal, NaBH(OAc)3, CH2Cl2, 4 Å MS; G: (i) TFA, CH2Cl2; (ii) Cs2CO3, DMF, Δ; H: 2-Br-C6H4CHO, NaBH(OAc)3, CH2Cl2, 4 Å MS; I: 5 mol% Pd(PPh3)4, Et3N, MeCN, μw, 125 °C; J: allyl bromide, K2CO3, DMF; K: 2.5 mol% Grubbs II, TsOH, PhMe, Δ; L: EDCI, N-Boc-glycine, Et3N, CH2Cl2. a 15 MeOH–PhMe, 65 °C; F: N-Boc-glycinal, NaBH(OAc)3, CH2Cl2, 4 Å MS; G: (i) TFA, CH2Cl2; (ii) Cs2CO3, DMF, Δ; H: 2-Br-C6H4CHO, NaBH(OAc)3, CH2Cl2, 4 Å MS; I: 5 mol% Pd(PPh3)4, Et3N, MeCN, μw, 125 °C; J: allyl bromide, K2CO3, DMF; K: 2.5 mol% Grubbs II, TsOH, PhMe, Δ; L: EDCI, N-Boc-glycine, Et3N, CH2Cl2. a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) See ESI† for variation from standard methods. b See ESI† for variation from standard methods. b![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Product not purified before subsequent reaction. c Product not purified before subsequent reaction. c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Yield over 2 steps. d Yield over 2 steps. d![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 92 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 mixture with the regioisomeric azocane. e 8 mixture with the regioisomeric azocane. e![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) The regioisomeric azocane 14c also isolated (31% yield). The regioisomeric azocane 14c also isolated (31% yield). | ||
1,2-Amino alcohols, 1,2-diamines and their functionalised derivatives are prevalent in many bioactive molecules.10 We therefore explored introduction of these motifs by pairwise coupling of the π-nucleophilic allyl group with the nucleophilic N-acyl substituents of 1 and 6 (carbamate11 or urea12 respectively) upon activation with iodine; subsequent treatment with NaN3 gave cyclic carbamates 5a–5d and the biologically relevant13 ureas 7a–c.
Pairwise coupling of the electrophilic carboxylate ester with nitrogen-centred nucleophiles led to hydantoins 8a,8c–d (a motif known for anticonvulsant biological activity, e.g. phenytoin) from acyl ureas 6 and lactams 10a–b/diketopiperazine 11d from protected amines 9. Finally, ring closure between the N-substituent and the C2-allyl group could be facilitated through transition metal catalysis: Pd-catalysed Heck cyclisations of substrates 12 gave the azepanes 13a–d and the azocane 14c. Similarly, Ru-catalysed ring-closing metathesis allowed the synthesis of the indolizidine alkaloid-like tetrahydropyridines 16a and the congeners 16b–d. In this way we prepared 22 distinct scaffolds from the four building blocks 1a–d in a total of just 49 synthetic operations‡ using a toolkit consisting of only six reaction methodologies and one N-capping event.
To assess the novelty of these scaffolds, substructure searches against the ZINC and CAS databases were performed. A total of 37 variants of the scaffolds (with protecting groups removed and allowing for modifications available through standard functional group interconversions; see ESI† for full details) were screened against the ZINC database of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 039
039![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 756 commercially available compounds. This returned only 28 hits, all of which were derived from just two of the scaffolds (10a/16a). A further analysis of the Murcko assemblies of all scaffolds (with alpha-attachments) against a random 5% of the ZINC database again only returned hits (4) against the same two scaffolds 10a/16a. Thus, it is clear that the scaffolds display a very high degree of novelty relative to commercial collections. Finally, the scaffolds were also assessed for skeletal diversity: using an hierarchical approach14,15 it was found that each scaffold comprised a unique molecular framework at the GNB level.§ Thus, the scaffolds are not simple derivatives of each other, but represent a skeletally diverse collection.
756 commercially available compounds. This returned only 28 hits, all of which were derived from just two of the scaffolds (10a/16a). A further analysis of the Murcko assemblies of all scaffolds (with alpha-attachments) against a random 5% of the ZINC database again only returned hits (4) against the same two scaffolds 10a/16a. Thus, it is clear that the scaffolds display a very high degree of novelty relative to commercial collections. Finally, the scaffolds were also assessed for skeletal diversity: using an hierarchical approach14,15 it was found that each scaffold comprised a unique molecular framework at the GNB level.§ Thus, the scaffolds are not simple derivatives of each other, but represent a skeletally diverse collection.
The potential of the scaffolds to provide access to lead-like screening compounds was demonstrated by enumeration of a virtual library of 1110 compounds from the deprotected scaffolds and 80 typical medicinal chemistry capping groups (see ESI†). Assessment of the lead-likeness of the virtual library (Fig. 1, Panel A) showed that 66% of compounds survived filtering by molecular size (14 ≤ heavy atom count ≤ 26; μ = 22.8, σ = 3.57), lipophilicity (−1 < A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 3; μ = 0.38, σ = 1.38) and structural filters (see ESI†). By comparison, just 23% of the ∼9 M compounds from the ZINC database15 survived this filtering process, with most compounds lying well outside lead-like chemical space (heavy atoms: μ = 25.9, σ = 5.4; A
P < 3; μ = 0.38, σ = 1.38) and structural filters (see ESI†). By comparison, just 23% of the ∼9 M compounds from the ZINC database15 survived this filtering process, with most compounds lying well outside lead-like chemical space (heavy atoms: μ = 25.9, σ = 5.4; A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P: μ = 1.7, σ = 2.9). Remarkably, using the same set of capping groups, each of the 22 scaffolds allowed significant lead-like chemical space to be targeted. In addition to this in silico study, we have shown practically that decoration of exemplar scaffolds is possible (ESI†).
P: μ = 1.7, σ = 2.9). Remarkably, using the same set of capping groups, each of the 22 scaffolds allowed significant lead-like chemical space to be targeted. In addition to this in silico study, we have shown practically that decoration of exemplar scaffolds is possible (ESI†).
The shape diversity of the virtual library was compared with that of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 911 randomly-selected compounds from the ZINC database (Fig. 1, Panel B). For each compound, the two normalised principal moments of inertia values were determined for a low energy conformation.16 The virtual library was significantly more three-dimensional (Fsp3: μ = 0.57) than the commercially-available compounds (Fsp3: μ = 0.33).
911 randomly-selected compounds from the ZINC database (Fig. 1, Panel B). For each compound, the two normalised principal moments of inertia values were determined for a low energy conformation.16 The virtual library was significantly more three-dimensional (Fsp3: μ = 0.57) than the commercially-available compounds (Fsp3: μ = 0.33).
In this study we have shown that careful selection of small, polyfunctional substrates facilitates an efficient approach to the synthesis of molecular scaffolds that are novel, diverse, and can specifically target lead-like chemical space. This was achieved using a toolkit of just six cyclisation methodologies, leading to 22 scaffolds in only 49 synthetic operations. In particular, the efficient and diverse coverage of 3D space from such a small set of reagents is notable. This general approach should be applicable to many classes of polyfunctional substrate, enabling the more efficient exploration of lead-like chemical space.
We thank EPSRC (EP/J00894X) and GSK for funding.
Notes and references
-
(a) M. C. Wenlock, R. P. Austin, P. Barton, A. M. Davis and P. D. Leeson, J. Med. Chem., 2003, 46, 1250 CrossRef CAS PubMed
; (b) P. D. Leeson and B. Springthorpe, Nat. Rev. Drug Discovery, 2007, 6, 881 CrossRef CAS PubMed
.
- F. Lovering, J. Bikker and C. Humblet, J. Med. Chem., 2009, 52, 6752 CrossRef CAS PubMed
.
-
(a) T. I. Oprea, A. M. Davis, S. J. Teague and P. D. Leeson, J. Chem. Inf. Comput. Sci., 2001, 41, 1308 CrossRef CAS PubMed
; (b) G. M. Keserü and G. M. Makara, Nat. Rev. Drug Discovery, 2009, 8, 203 CrossRef PubMed
.
- A. Nadin, C. Hattotuwagama and I. Churcher, Angew. Chem., Int. Ed., 2012, 51, 1114 CrossRef CAS PubMed
.
- A. H. Lipkus, Q. Yuan, K. A. Lucas, S. A. Funk, W. F. Bartelt, R. J. Schenck and A. J. Trippe, J. Org. Chem., 2008, 73, 4443 CrossRef CAS PubMed
.
- P. MacLellan and A. Nelson, Chem. Commun., 2013, 49, 2383 RSC
.
- R. Doveston, S. Marsden and A. Nelson, Drug Discovery Today, 2014, 19, 813 CrossRef CAS PubMed
.
-
(a) R. G. Doveston, P. Tosatti, M. Dow, D. J. Foley, H. Y. Li, A. J. Campbell, D. House, I. Churcher, S. P. Marsden and A. Nelson, Org. Biomol. Chem., 2015, 13, 859 RSC
; (b) S. V. Ryabukhin, D. M. Panov, D. S. Granat, E. N. Ostapchuk, D. V. Kryvoruchko and O. O. Grygorenko, ACS Comb. Sci., 2014, 16, 146 CrossRef CAS PubMed
; (c) A. Borisov, V. Voloshchuk, M. Nechayev and O. Grygorenko, Synthesis, 2013, 2413 CAS
; (d) T. James, I. Simpson, J. A. Grant, V. Sridharan and A. Nelson, Org. Lett., 2013, 15, 6094 CrossRef CAS PubMed
; (e) T. James, P. MacLellan, G. M. Burslem, I. Simpson, J. A. Grant, S. Warriner, V. Sridharan and A. Nelson, Org. Biomol. Chem., 2014, 12, 2584 RSC
.
- P. N. Confalone, E. M. Huie, S. S. Ko and G. M. Cole, J. Org. Chem., 1988, 53, 482 CrossRef CAS
.
-
(a) T. J. Donohoe, C. K. A. Callens, A. Flores, A. R. Lacy and A. H. Rathi, Chem. – Eur. J., 2011, 17, 58 CrossRef CAS PubMed
; (b) D. Lucet, T. Le Gall and C. Mioskowski, Angew. Chem., Int. Ed., 1998, 37, 2580 CrossRef CAS
.
- M. Pattarozzi, C. Zonta, Q. B. Broxterman, B. Kaptein, R. De Zorzi, L. Randaccio, P. Scrimin and G. Licini, Org. Lett., 2007, 9, 2365 CrossRef CAS PubMed
.
- M. Fujita, O. Kitagawa, T. Suzuki and T. Taguchi, J. Org. Chem., 1997, 62, 7330 CrossRef CAS PubMed
.
- E. De Clercq, Biochim. Biophys. Acta, 2002, 1587, 258 CrossRef CAS
.
- A. Schuffenhauer, P. Ertl, S. Roggo, S. Wetzel, M. A. Koch and H. Waldmann, J. Chem. Inf. Model., 2007, 47, 47 CrossRef CAS PubMed
.
- J. J. Irwin, T. Sterling, M. M. Mysinger, E. S. Bolstad and R. G. Coleman, J. Chem. Inf. Model., 2012, 52, 1757 CrossRef CAS PubMed
.
- W. H. Sauer and M. K. Schwarz, J. Chem. Inf. Comput. Sci., 2003, 43, 987 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, compound characterisation data, copies of 1H and 13C NMR spectra, and details of the virtual library enumeration/analysis. See DOI: 10.1039/c5cc03002a |
| ‡ Defined as a process conducted in a single reaction vessel. |
| § See the scaffold tree in ESI† for further detail. |
| This journal is © The Royal Society of Chemistry 2015 |