Open Access Article
Open Access ArticleSaccharothriolides A–C, novel phenyl-substituted 10-membered macrolides isolated from a rare actinomycete Saccharothrix sp.†
Shan
Lu
a,
Shinichi
Nishimura
a,
Go
Hirai
b,
Masashi
Ito
c,
Teppei
Kawahara
d,
Miho
Izumikawa
d,
Mikiko
Sodeoka
b,
Kazuo
Shin-ya
e,
Toshio
Tsuchida
c and
Hideaki
Kakeya
*a
aDepartment of System Chemotherapy and Molecular Sciences, Division of Bioinformatics and Chemical Genomics, Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: scseigyo-hisyo@pharm.kyoto-u.ac.jp
bSynthetic Organic Chemistry Laboratory, RIKEN, Wako, Saitama 351-0198, Japan
cResearch & Development Division, MicroBioPharm Japan Co., Ltd. (MBJ), Iwata, Shizuoka 438-0078, Japan
dJapan Biological Informatics Consortium (JBIC), Koto-ku, Tokyo 135-0064, Japan
eNational Institute of Advanced Industrial Science and Technology (AIST), Koto-ku, Tokyo 135-0064, Japan
First published on 1st April 2015
Abstract
Three new 10-membered macrolides, saccharothriolides A–C (1–3), were discovered from a rare actinomycete Saccharothrix sp. A1506. All of the sp3 carbons in the 10-membered ring had chirality, which was determined by extensive spectroscopic analysis and TDDFT-calculation of ECD spectra. Saccharothriolide B (2) exhibited cytotoxicity against human tumor cell lines HeLa and HT1080.
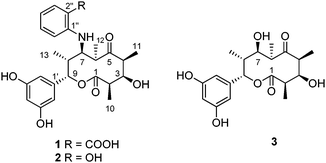
Rare actinomycetes (or non-streptomycete actinomycetes) are regarded as the actinomycete strains whose isolation frequency determined by conventional methods is much lower than that of the streptomycete strains. These under-explored microorganisms are expected to be a rich source of natural products with novel chemical structures and biological activities.1,2Saccharothrix sp., one of the rare actinomycetes, was first obtained from a soil sample collected in Australia in 1984.3 Dozens of bioactive natural products with a structural and biological diversity have been isolated from this genus: for example, a naphthoquinone derivative sacchathridine A which was reported as a prostaglandin release inhibitor,4 16-membered macrolides tianchimycins A and B,5 cytotoxic 20-membered macrolides ammocidins A–D,6,7 antibacterial galacardins A and B,8 and antitumor rebeccamycin.9 In the course of our chemical screening for novel microbe metabolites, we discovered new 10-membered macrolides from a rare actinomycete Saccharothrix sp. A1506, designated as saccharothriolides A–C (1–3) (Fig. 1). Here we report their isolation, structure elucidation, and biological activities.
We surveyed more than 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 microbe cultures by LC-MS analysis to identify a novel metabolite from a culture broth of a rare actinomycete Saccharothrix sp. A1506. This metabolite, named saccharothriolide A (1), exhibited an MS signal (m/z 486.2134 [M + H]+) which was not found in metabolites database and a UV absorption at 345 nm characteristic to anthranilic acid. Further analysis of the LC-MS data revealed the presence of two congeners, saccharothriolides B (2) and C (3). By LC-MS-guided isolation, we purified metabolites 1 (24.7 mg), 2 (5.4 mg), and 3 (17.8 mg) from a 6 L-culture.
000 microbe cultures by LC-MS analysis to identify a novel metabolite from a culture broth of a rare actinomycete Saccharothrix sp. A1506. This metabolite, named saccharothriolide A (1), exhibited an MS signal (m/z 486.2134 [M + H]+) which was not found in metabolites database and a UV absorption at 345 nm characteristic to anthranilic acid. Further analysis of the LC-MS data revealed the presence of two congeners, saccharothriolides B (2) and C (3). By LC-MS-guided isolation, we purified metabolites 1 (24.7 mg), 2 (5.4 mg), and 3 (17.8 mg) from a 6 L-culture.
Saccharothriolide A (1) was obtained as a light yellow oil with [α]20D + 18.0 (c = 0.74, MeOH). The molecular formula was determined to be C26H31NO8 by HR-ESI-MS (m/z 486.2134 [M + H]+, calcd 486.2128), indicating the presence of 12 degrees of unsaturation. The IR spectrum showed absorptions corresponding to hydroxyl (3327 cm−1), ester carbonyl (1732 cm−1), and ketone (1679 cm−1) groups. The 1H NMR spectrum of 1 displayed seven aromatic protons (δH 7.95, 7.31, 6.66, 6.57, 6.06, 5.87 × 2 ppm) and two oxymethine protons (δH 5.50, 3.79 ppm) in addition to four aliphatic methyl signals (δH 1.09, 1.20, 1.35, 1.46 ppm) and five methine protons (δH 2.19, 2.88, 3.28, 3.43, 3.66 ppm) (Table 1). The 13C NMR spectrum included 26 carbon signals, corresponding to four aliphatic methyls (δC 10.7, 14.1, 18.4, 20.3), seven aliphatic methines including two carbons adjacent to an oxygen atom (δC 74.3 and 81.4) and one carbon adjacent to a nitrogen atom (δC 62.2), twelve aromatic carbons and three carbonyl carbons (δC 173.8 × 2, 224.6).
| No. | 1 | |
|---|---|---|
| δ H, m, J (Hz) | δ C | |
| 1 | — | 173.8, C |
| 2 | 2.88, qd, 6.9, 3.4 | 46.3, CH |
| 3 | 3.79, brs | 81.4, CH |
| 4 | 3.28, q, 7.5 | 50.8, CH |
| 5 | — | 224.6, C |
| 6 | 3.43, q, 6.9 | 43.7, CH |
| 7 | 3.66, brs | 62.2, CH |
| 8 | 2.19, qd, 6.9, 5.2 | 43.1, CH |
| 9 | 5.50, brs | 74.3, CH |
| 10 | 1.20, d, 6.9 | 14.1, CH3 |
| 11 | 1.46, d, 7.5 | 18.4, CH3 |
| 12 | 1.35, d, 6.9 | 20.3, CH3 |
| 13 | 1.09, d, 6.9 | 10.7, CH3 |
| 1′ | — | 144.9, C |
| 2′,6′ | 5.87, s | 104.7, CH |
| 3′,5′ | — | 159.5, C |
| 4′ | 6.06, s | 102.3, CH |
| 1′′ | — | 151.7, C |
| 2′′ | — | 113.7, C |
| 3′′ | 7.95, brs | 134.1, CH |
| 4′′ | 6.57, t, 5.7 | 115.6, CH |
| 5′′ | 7.31, t, 7.5 | 135.6, CH |
| 6′′ | 6.66, d, 8.6 | 111.8, CH |
| 2′′-COOH | — | 173.8, C |
The 1H–1H COSY experiment revealed the presence of three spin systems: CH3-10/H-2/H-3/H-4/CH3-11, CH3-12/H-6/H-7/H-8/CH3-13/H-9, and H-3′′/H-4′′/H-5′′/H-6′′ (Fig. 2). HMBC correlations from CH3-11 to C-5, and from CH3-12 to C-5 connected C-4 and C-6 through a ketone group. HMBC correlations from both H-3 and H-9 to carbonyl C-1 connected C-2 and C-9 through an ester bond, leading to the formation of the 10-membered lactone ring (Fig. 2). The formation of the lactone ring was further supported by the observation of the down-field shifted chemical shift for H-9 (δH 5.50). The meta-disubstituted benzene ring was determined on the basis of the HMBC correlations from H-4′ to C-2′/C-6′, and from H-2′/H-6′ to C-3′/C-5′. This benzene ring was connected to C-9 due to the HMBC correlations from H-9 to aromatic carbons C-1′ and C-2′/6′. The presence of anthranilic acid suggested by the characteristic UV absorption (Fig. S6, ESI†)10 was confirmed by HMBC correlations from H-6′′ (δH 6.66) to C-2′′ (δC 113.7), and from H-3′′ (δH 7.95) to C-1′′ (δC 151.7) and 2′′-COOH (δC 173.8), along with the 1H–1H COSY correlations from H-3′′ to H-6′′ (Fig. 2). The anthranilic acid was connected to the lactone ring at C-7 through an NH group, which was deduced by the up-field shifted chemical shift for C-7 (δC 62.2) and the presence of a free carboxylic acid (δC 173.8). This connection was further confirmed by the mutual NOESY correlations among H-6′′, H-7, and H-8 (Fig. 2).
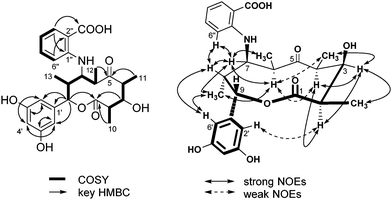 | ||
| Fig. 2 1H–1H COSY (left, bold) correlations and selected HMBC (left, arrow) and NOESY (right) correlations in saccharothriolide A (1). | ||
The relative stereochemistry of metabolite 1 was deduced from the NOESY data (Fig. 2). NOESY cross peaks between H-2 and H-4 indicated that they are placed in the same α face, whereas NOESY correlations between H-3 and H-2, H-4, CH3-10, and CH3-11 suggested α configuration for H-3. NOESY correlations between H-6 and H-4(weak), CH3-11(weak), and CH3-13 indicated an α configuration for these protons. A β configuration for H-8 was then determined. Correlations between H-7 and H-6, H-8, CH3-12, and CH3-13 suggested that H-7 has an α configuration. Finally, NOESY correlations between the aromatic proton H-6′ and H-8, and CH3-13, together with a weak correlation from the aromatic proton H-2′ to H-2, revealed the β orientation of H-9. Thus, the relative configurations were deduced to be 2R*, 3R*, 4S*, 6R*, 7R*, 8R*, 9S*.
Macrolides can exist in several conformations, which can be a potential cause of mis-interpretation of the NOESY data. We next analyzed the relative stereochemistry of 1 by the advanced statistical Universal NMR Database (UDB) approach, originally developed by Kishi and co-workers.11–13 Reductive opening of the lactone ring of 1 using LiAlH4 furnished a linear product 4 (Fig. 3). The 13C NMR data of the two tetrad segments, C3–C6 and C5–C2, were subjected to the statistical UDB analysis. The difference between the adjusted NMR data of the tetrad segments in 4 and Kishi's database was calculated to reveal that both sequences have an anti–anti–anti configuration (Table S2–S5, ESI†), which was in good agreement with the NOESY data in 1.
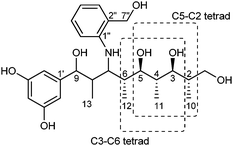 | ||
| Fig. 3 Analysis of the stereochemistry of compound 4 (C3–C6 and C5–C2 tetrads) by the statistical UDB approach. | ||
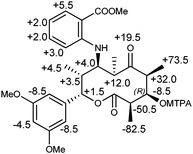
The absolute stereochemistry was determined by the modified Mosher's method (Fig. 4).14 Phenolic hydroxy groups and carboxylic acid of 1 were first protected by methylation using CH3I to yield a tri-methyl derivative 5. The methylated derivative 5 was treated with (R)- and (S)-MTPA chloride to afford (S)- and (R)-MTPA esters of 5, respectively. The Δδ (δS–δR) values of the protons flanking the C-3 chiral center revealed the 3R absolute configuration, which in turn concluded the absolute configurations of 1 as 2R, 3R, 4S, 6R, 7R, 8R, 9S.
The modified Mosher's method is sometimes not applicable to axial hydroxyl groups.15 In order to confirm the above results, we finally measured the CD spectrum of 1, which was compared with the electronic circular dichroism (ECD) spectrum calculated by the time-dependent density functional theory (TDDFT).16 As shown in Fig. 5, the experimental CD spectrum showed a Cotton effect at 199 (Δε, +15.0), 227 (Δε, −25.0) and 355 (Δε, +3.46) nm, all of which were observed in the ECD spectrum calculated for the stereochemistry of 2R, 3R, 4S, 6R, 7R, 8R, 9S. Thus, the absolute configuration of 1 was unambiguously established by two independent methods.
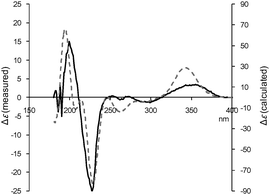 | ||
| Fig. 5 Experimental CD (solid line) and calculated ECD (dotted line) spectra of saccharothriolide A (1). ECD was calculated for 2R, 3R, 4S, 6R, 7R, 8R, 9S. | ||
To find congeners of saccharothriolide A (1), we investigated the LC-MS data of the crude broth extract. We detected a metabolite whose ion peak was observed at m/z 458.2162 ([M + H]+), revealing the molecular formula of C25H31NO7. This metabolite, designated as saccharothriolide B (2), contained one nitrogen atom similar to 1, but its molecular size was 28 Da smaller than 1. Metabolite 2 was obtained as a light yellow oil with [α]20D −81.2 (c = 0.36, MeOH).
The 1H and 13C NMR data of 2 were very similar to those of 1 (Table S1, ESI†), with the exception of the disappearance of the carboxylic acid. The obvious downfield shift of C-2′′ (δC 113.7 in 1, δC 146.0 in 2) and an upfield shift of C-1′′ and C-3′′ (δC 151.7 in 1, δC 138.2 in 2 and δC 134.1 in 1, δC 115.2 in 2, respectively) were observed, accompanied by a marked upfield shift of H-3′′ and H-5′′ (δH 7.95 in 1, δH 6.73 in 2 and δH 7.31 in 1, δH 6.70 in 2, respectively), which suggested that 2 possessed a phenolic hydroxyl group instead of a carboxylic acid at C-2′′. This was in agreement with the fact that 2 was 28 Da smaller than 1. The planar structure was deduced by the COSY, HMQC and HMBC data (Fig. S1, ESI†). Detailed analysis of the NOESY spectrum of 2 revealed that the relative stereochemistry was the same as that of 1 (Fig. S1, ESI†): 2R*, 3R*, 4S*, 6R*, 7R*, 8R*, 9S*.
The absolute stereochemistry of 2 was determined by measurement and calculation of ECD. The ECD calculation of 2 revealed that the absolute stereochemistry was the same as that of 1. The calculated ECD spectrum overlapped well with the experimental CD spectrum (Fig. 6), concluding that the absolute stereochemistry of 2 was 2R, 3R, 4S, 6R, 7R, 8R, 9S.
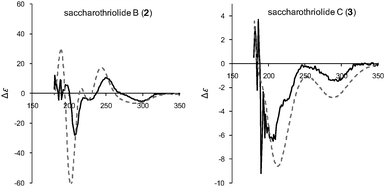 | ||
| Fig. 6 Experimental CD (solid line) and calculated ECD (dotted line) spectra for saccharothriolides B (2) and C (3). Both ECD spectra were calculated for 2R, 3R, 4S, 6R, 7R, 8R, 9S. | ||
Saccharothriolides A (1) and B (2) have an aminoaryl substitution at C-7, suggesting the presence of a common precursor for them, i.e., Michael addition of nucleophilic amines to the corresponding precursor can afford metabolites 1 and 2. For investigating this possibility, we surveyed the LC-MS data of the culture broth to identify saccharothriolide C (3). Metabolite C (3) was obtained as a light yellow oil with [α]20D −111.8 (c = 0.58, MeOH). The HR-ESI-MS data indicated an [M + Na]+ ion peak at m/z 389.1586 ([M + Na]+, calcd 389.1576), revealing a molecular formula of C19H26O7. The 1H and 13C NMR data of 3 were similar to those of 1 and 2 (Table S1, ESI†), except for the absence of a set of aromatic protons and carbon signals corresponding to the amino aryl groups substituted at C-7. Instead, metabolite 3 possessed a hydroxyl group at C-7, supported by the downfield shift of the carbon signal of C-7 (δC 80.1). The planar structure was elucidated by detailed analysis of the 2D NMR data, while the NOESY data indicated the same relative configuration as those of 1 and 2 (Fig. S2, ESI†). As expected, the absolute stereochemistry of 3 was the same as those of 1 and 2, because the calculated ECD spectrum for the stereochemistry of 2R, 3R, 4S, 6R, 7R, 8R, 9S showed a high similarity to that of the measured one (Fig. 6).
Metabolite 2 exhibited moderate cytotoxicity against cancer cells including HeLa and HT1080 cell lines with IC50 values of 17.9 and 13.9 μM, respectively. Other saccharothriolides 1 and 3 were inactive even at 100 μM. Additionally, only metabolite 2 showed weak antibacterial activity at 50 μg per disc against Staphylococcus aureus in a paper disc assay. These results indicate that saccharothriolides are capable of regulating their biological activities by modifying the functional group at the C-7 position.
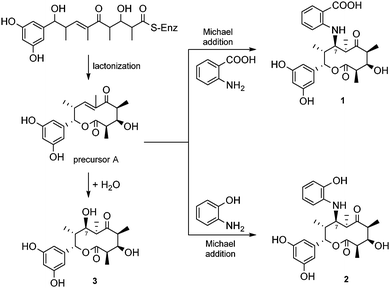
Saccharothriolides are 10-membered macrolides. As in the case of other macrolides, saccharothriolides seem to be synthesized via the polyketide biosynthetic pathway (Scheme 1). An aryl starter unit and four units of methyl-malonyl-CoA seem to be conjugated followed by cyclization to yield the precursor metabolite A. The aromatic unit-priming polyketide system has been found only in a limited number of metabolites, i.e. soraphen A from Sorangium cellulosum,17 rifamycins from Amycolatopsis rifamycinica18 and enterocin from Streptomyces maritimus.19 The precursor A is likely attacked by an aminoaryl group to furnish metabolites 1 and 2, or by a water molecule to furnish metabolite 3 (Scheme 1). In fact, the culture broth included a significant amount of anthranilic acid (23.5 mg) and 2-hydroxyacetanilide (120.9 mg).20 Although we explored the presence of precursor A using the LC-MS data, ion peaks corresponding to the precursor were not found, probably due to the high reactivity of the α,β-unsaturated ketone.
In conclusion, we discovered three novel 10-membered macrolides from a rare actinomycete Saccharothrix sp. A1506. Structural analysis implied their common biosynthetic origin, whereas we predict the presence of precursor A possessing α,β-unsaturated ketone. It should be noted that only saccharothriolide B (2) showed moderate biological activities. Detailed studies on the biosynthetic mechanism and modes of action are currently being undertaken in our laboratory.
We thank the RIKEN Integrated Cluster of Clusters (RICC) at RIKEN for the computer resources used for the calculation. This work was supported in part by research grants from the Japan Society for the Promotion of Science (JSPS), the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and the Ministry of Health, Labour and Welfare of Japan (MHLW).
Notes and references
- J. Bérdy, J. Antibiot., 2005, 58, 1–26 CrossRef PubMed.
- K. Tiwari and R. K. Gupta, Crit. Rev. Biotechnol., 2012, 32, 108–132 CrossRef CAS PubMed.
- D. P. Labeda, R. T. Testa, M. P. Lechaevalier and H. A. Lechevalier, Int. J. Syst. Bacteriol., 1984, 34, 426–431 CrossRef.
- K. Nakae, I. Kurata, F. Kojima, M. Igarashi, M. Hatano, R. Sawa, Y. Kubota, H. Adachi and A. Nomoto, J. Nat. Prod., 2013, 76, 720–722 CrossRef CAS PubMed.
- X. L. Wang, J. Tabudravu, M. Jaspars and H. Deng, Tetrahedron, 2013, 69, 6060–6064 CrossRef CAS.
- R. Murakami, T. Tomikawa, K. Shin-ya, J. Shinozaki, T. Kajiura, T. Kinoshita, A. Miyajima, H. Seto and Y. Hayakawa, J. Antibiot., 2001, 54, 710–713 CrossRef CAS PubMed.
- R. Murakami, J. Shinozaki, T. Kajiura, I. Kozone, M. Takagi, K. Shin-Ya, H. Seto and Y. Hayakawa, J. Antibiot., 2009, 62, 123–127 CrossRef CAS PubMed.
- M. Takeuchi, S. Takahashi, R. Enokita, Y. Sakaida, H. Haruyama, T. Nakamura, T. Katayama and M. Inukai, J. Antibiot., 1992, 45, 297–305 CrossRef CAS PubMed.
- D. E. Nettleton, T. W. Doyle, B. Krishnan, G. K. Matsumoto and J. Clardy, Tetrahedron Lett., 1985, 26, 4011–4014 CrossRef CAS.
- T. Stalin and N. Rajendiran, J. Photochem. Photobiol., A, 2006, 182, 137–150 CrossRef CAS.
- E. Fleury, M. I. Lannou, O. Bistri, F. Sautel, G. Massiot, A. Pancrazi and J. Ardisson, Eur. J. Org. Chem., 2009, 4992–5001 CrossRef CAS.
- Y. Kobayashi, J. Lee, K. Tezuka and Y. Kishi, Org. Lett., 1999, 1, 2177–2180 CrossRef CAS PubMed.
- J. Lee, Y. Kobayashi, K. Tezuka and Y. Kishi, Org. Lett., 1999, 1, 2181–2184 CrossRef CAS PubMed.
- I. Ohtani, T. Kusumi, Y. Kashman and H. Kakisawa, J. Am. Chem. Soc., 1991, 113, 4092–4096 CrossRef CAS.
- J. M. Seco, E. Quinoá and R. Riguera, Chem. Rev., 2004, 104, 17–117 CrossRef CAS.
- We also calculated 3JH–H values for the stable conformer, which were almost the same as those of the measured ones (Table S6, ESI†).
- J. Ligon, S. Hill, J. Beck, R. Zirkle, I. Molnár, J. Zawodny, S. Money and T. Schupp, Gene., 2002, 285, 257–267 CrossRef CAS PubMed.
- P. R. August, L. Tang, Y. J. Yoon, S. Ning, R. Müller, T. W. Yu, M. Taylor, D. Hoffmann, C. G. Kim, X. H. Zhang, C. R. Hutchinson and H. G. Floss, Chem. Biol., 1998, 5, 69–79 CrossRef CAS PubMed.
- J. Piel, C. Hertweck, P. R. Shipley, D. M. Hunt, M. S. Newman and B. S. Moore, Chem. Biol., 2000, 7, 943–955 CrossRef CAS PubMed.
- Anthranilic acid and 2-hydroxyacetanilide were purified from a part of the 6 L-culture.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and compound data. See DOI: 10.1039/c5cc01953b |
| This journal is © The Royal Society of Chemistry 2015 |