Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films
Sachin K.
Srivastava
*ab,
Hilla Ben
Hamo
c,
Ariel
Kushmaro
bcd,
Robert S.
Marks
bcd,
Christoph
Grüner
e,
Bernd
Rauschenbach
ef and
Ibrahim
Abdulhalim
abd
aDepartment of Electro optic Engineering, Ben Gurion University of the Negev, Beer Sheva 84105, Israel. E-mail: sachinchitransh@gmail.com
bIlse Katz Institute for Nanoscale Science and Technology, Ben Gurion University of the Negev, Beer Sheva 84105, Israel
cThe Avram and Stella Goldstein-Goren Department of Biotechnology Engineering, Ben Gurion University, Beer Sheva-84105, Israel
dSchool of Material Science and Engineering, Nanyang Technological University, 637722, Singapore
eLeibniz Institute of Surface Modification, Permoserstrasse 15, 04318 Leipzig, Germany
fUniversity Leipzig, Institute for Experimental Physics II, Linnéstr. 5, 04307 Leipzig, Germany
First published on 25th February 2015
Abstract
A nanobiosensor chip, utilizing surface enhanced Raman spectroscopy (SERS) on nanosculptured thin films (nSTFs) of silver, was shown to detect Escherichia coli (E. coli) bacteria down to the concentration level of a single bacterium. The sensor utilizes highly enhanced plasmonic nSTFs of silver on a silicon platform for the enhancement of Raman bands as checked with adsorbed 4-aminothiophenol molecules. T-4 bacteriophages were immobilized on the aforementioned surface of the chip for the specific capture of target E. coli bacteria. To demonstrate that no significant non-specific immobilization of other bacteria occurs, three different, additional bacterial strains, Chromobacterium violaceum, Paracoccus denitrificans and Pseudomonas aeruginosa were used. Furthermore, experiments performed on an additional strain of E. coli to address the specificity and reusability of the sensor showed that the sensor operates for different strains of E. coli and is reusable. Time resolved phase contrast microscopy of the E. coli-T4 bacteriophage chip was performed to study its interaction with bacteria over time. Results showed that the present sensor performs a fast, accurate and stable detection of E. coli with ultra-small concentrations of bacteria down to the level of a single bacterium in 10 μl volume of the sample.
Introduction
Biosensing using surface enhanced Raman spectroscopy (SERS) has emerged as a popular field of research since the past decade due to its putative low detection limit, high sensitivity and specificity as well as other intrinsic exciting properties. A number of biosensors based on SERS for the detection of glucose, cocaine, DNA, endocrine disruption compounds (EDCs), E. coli, etc. have been reported.1–5 SERS is the phenomenon of enhancement of the Raman spectroscopy signal of certain molecules by a factor of several orders of magnitude when they are brought in contact with nanostructured metal surfaces. This enhancement in intensity is attributed to the highly localized fields of plasmons in the vicinity of the molecule emitting Raman signals, and many theoretical and experimental studies have been performed to elucidate its dependence on various factors6,7 (shape, size, orientation, porosity, material of the nanostructure and the substrates on which the nanostructures are deposited).In order to modulate signal enhancement a number of structures were studied, including nanorods, nanowires, nanocubes, dielectric–metallic core–shells, nanoflowers, nanosculptured thin films (nSTFs) and many other shapes of different material compositions and these were examined for SERS enhancement.8–12 nSTFs have unique properties such as large enhancement factor, stability, reproducibility, durability, ease of fabrication, large scale surfaces, cost effectiveness, etc. nSTFs are nanorod-like structures with different surface morphologies which are generally grown by the glancing angle deposition technique (GLAD).13 Previous studies by our group established the optimization of the performance of the SERS enhancement of nSTFs with respect to their material composition, height, underlying substrates and porosity4,12 and those determined to be optimal were used in the present study.
Escherichia coli is a Gram-negative, facultative anaerobic, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms. E. coli is considered to be an indicator for fecal contamination (fecal coliforms) and some strains are pathogenic, responsible for food or water borne, gastrointestinal diseases. Consumption of water and/or food contaminated with E. coli can lead to hemolytic-uremic syndrome (HUS), especially in children and elderly people. HUS causes the destruction of red blood cells and kidney failure, which may lead to stroke, seizures, and even death.14 According to surveys of the World Health Organization (WHO), approximately two billion people get affected by gastrointestinal diseases annually.5 According to the statistics of the United States Centres for Disease Control and Prevention (CDC), about 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people are affected annually by E. coli in the US alone.15 Therefore, E. coli has become a prime target for detection and cure. A number of classical methods (plate counting, polymerase chain reaction (PCR), ELISA), as well as, state-of-the-art methodologies including physical transduction methods such as surface plasmon resonance (SPR), long period fiber gratings (LPG), surface acoustic waves, surface enhanced fluorescence (SEF), microelectronic mechanical systems (MEMS), amperometric detection, microfluidics integrated microscopy, surface enhanced Raman spectroscopy (SERS), fiber optic immunosensors, etc. have been reported for the detection of E. coli.16–26 SERS has been found to provide the lowest limits of detection from orders of magnitude in its enhancement.27 Since the Raman bands of a chemical/biochemical species are unique, the enhanced Raman spectroscopy is considered one of the most accurate methods for detection. However, since the Raman enhancement is a very short range phenomenon, the complete Raman bands of only very small species can be assessed by SERS.4 The complete information of all the Raman bands from bigger species such as bacteria is thus inaccessible. Most of the studies based on SERS based detection of E. coli bacteria are based on spectral differentiation at high concentrations and are susceptible to interferences from the culture medium.28 There is still only limited literature available which discusses the sensitivity of such sensors. Such a study was recently reported with nSTFs on filter papers.28 However, the limit of detection of these sensors remains quite high. Furthermore, specificity of a sensor requires the use of affinity-based capture recognition elements such as antibodies, nucleic acids (DNA/RNA), aptamers, bacteriophages, glycoproteins, etc.5,21,29–32 Antibody-based sensors suffer from the possibility of cross-linking to unrelated bacteria exhibiting similar molecular structures, thus, resulting in false signals.33 Aptamer based sensors are particularly suited for hapten-sized target molecules but it is still of limited use in large entity capture such as that of bacteria. In addition, neither antibodies nor aptamers enable the discrimination between viable and non-viable cells.34 Bacteriophage-based sensors exhibit high specificity (for their host bacteria) and relatively better stability than antibodies (from heat, alkali and acidic solvents).32 However, unlike antibodies and aptamers they need to be isolated from nature; however, once this is done they are easily cultured. Their specificity can be modulated in the laboratory through complicated laboratory-based mutation studies. Apart from that, the bacteriophages can easily discriminate living bacteria from dead ones as they do not infect dead cells. During interaction with living cells the bacteriophages inject their DNA into them, thus prompting the production of phages and subsequent lysis of the host cells.22 After such an interaction the phages cannot be used to recognize the host bacteria again. However, the sensor can be reused if there has been no specific recognition and therefore the phages are still intact.23
000 people are affected annually by E. coli in the US alone.15 Therefore, E. coli has become a prime target for detection and cure. A number of classical methods (plate counting, polymerase chain reaction (PCR), ELISA), as well as, state-of-the-art methodologies including physical transduction methods such as surface plasmon resonance (SPR), long period fiber gratings (LPG), surface acoustic waves, surface enhanced fluorescence (SEF), microelectronic mechanical systems (MEMS), amperometric detection, microfluidics integrated microscopy, surface enhanced Raman spectroscopy (SERS), fiber optic immunosensors, etc. have been reported for the detection of E. coli.16–26 SERS has been found to provide the lowest limits of detection from orders of magnitude in its enhancement.27 Since the Raman bands of a chemical/biochemical species are unique, the enhanced Raman spectroscopy is considered one of the most accurate methods for detection. However, since the Raman enhancement is a very short range phenomenon, the complete Raman bands of only very small species can be assessed by SERS.4 The complete information of all the Raman bands from bigger species such as bacteria is thus inaccessible. Most of the studies based on SERS based detection of E. coli bacteria are based on spectral differentiation at high concentrations and are susceptible to interferences from the culture medium.28 There is still only limited literature available which discusses the sensitivity of such sensors. Such a study was recently reported with nSTFs on filter papers.28 However, the limit of detection of these sensors remains quite high. Furthermore, specificity of a sensor requires the use of affinity-based capture recognition elements such as antibodies, nucleic acids (DNA/RNA), aptamers, bacteriophages, glycoproteins, etc.5,21,29–32 Antibody-based sensors suffer from the possibility of cross-linking to unrelated bacteria exhibiting similar molecular structures, thus, resulting in false signals.33 Aptamer based sensors are particularly suited for hapten-sized target molecules but it is still of limited use in large entity capture such as that of bacteria. In addition, neither antibodies nor aptamers enable the discrimination between viable and non-viable cells.34 Bacteriophage-based sensors exhibit high specificity (for their host bacteria) and relatively better stability than antibodies (from heat, alkali and acidic solvents).32 However, unlike antibodies and aptamers they need to be isolated from nature; however, once this is done they are easily cultured. Their specificity can be modulated in the laboratory through complicated laboratory-based mutation studies. Apart from that, the bacteriophages can easily discriminate living bacteria from dead ones as they do not infect dead cells. During interaction with living cells the bacteriophages inject their DNA into them, thus prompting the production of phages and subsequent lysis of the host cells.22 After such an interaction the phages cannot be used to recognize the host bacteria again. However, the sensor can be reused if there has been no specific recognition and therefore the phages are still intact.23
In the present study, we have fabricated chips of silver nSTFs over a Si substrate for the development of the sensor. SERS signals from model 4-aminothiophenol (4-ATP) were assessed for their sensing power. The specificity of the sensor was given by functionalization with T4 bacteriophages as the capture biomolecular recognition element. SERS spectra were recorded for two different strains (E. coli B and E. coli μX) at different concentrations, as well as, three unrelated control bacteria, namely Chromobacterium violaceum, Paracoccus denitrificans and Pseudomonas aeruginosa to confirm the fact that our sensor does not suffer from non-specific binding to the surface. Another control experiment was performed to observe the interaction/possibilities of lysis of E. coli on the sensor surface by developing the sensor protocol on a glass micro-slide. The binding/interaction of the attached phage – E. coli was recorded by a phase contrast microscope at different time intervals.
Materials and methods
4-Aminothiophenol (422967), glutaraldehyde (G7651), bovine serum albumin (BSA) (A2153), and (3-aminopropyl) trimethoxysilane (281778) (aminosilane) were purchased from Sigma. Distilled water of 18 MΩ cm resistivity was obtained from a Millipore® system. Phosphate buffer saline (PBS) was obtained from Dulbecco. Sulphuric acid (H2SO4, 98% pure) (19550501), hydrogen peroxide (H2O2, 30%) (08550323), ethanol (C2H5OH, dehydrated, 99.9% pure) (05250502), and acetic acid (CH3COOH, 99.8% pure) (01070521), were purchased from Bio-Lab Ltd, Israel. All the chemicals were used as obtained without further purification.Bacterial growth and culture
Frozen stocks of bacterial strains E. coli RFM44335 (E. coli B, ATCC® 11303™), E. coli XLMRF (E. coli μX), Paracoccus denitrificans (P. denitrificans), Pseudomonas aeruginosa (P. aeruginosa) and Chromobacterium violaceum 026 (C. violaceum)., a gift from Prof. P. Williams (University of Nottingham, Nottingham, UK), were used as seeds for cultivation in 10 ml LB (Difco Luria–Bertani medium, BD, France) grown overnight at 37 °C and 30 °C respectively in a rotary thermo-shaker (Gerhardt, Germany) at 120 rpm. The overnight culture was centrifuged for 10 min at 3000g, the pellet washed and re-suspended in 0.01 M phosphate-buffered saline (PBS), pH 7.2. The bacterial concentrations were determined using the colony forming units (CFU) method. C. violaceum is also a Gram-negative bacterium, but evolutionarily distant enough from E. coli, and cannot be recognized by T4 bacteriophage. Similar to E. coli, C. violaceum is commonly found both in soil and aquatic environments, but is not present as part of the normal flora of humans and animals.36,37P. aeruginosa, an opportunistic pathogen, is also found in water, soil, skin flora and man-made environments and is commonly involved in drinking water contamination events, while P. denitrificans is a soil bacterium.38–40 They both are Gram-negative bacteria.T4 bacteriophage preparation
Phage propagation was done as described before.23 A T4 phage culture was incubated with an E. coli broth, then added to fresh LB media and incubated as described before for 6 hours. The culture was centrifuged and the supernatant filtered. The phage suspension was centrifuged inside an Amicon® Ultra-0.5 (MILLIPORE) and the resulting phage pellet resuspended in SM buffer.41 Using the soft agar overlay technique the phage was enumerated by plaque forming unit (PFU).n-STF fabrication
Nanosculptured thin films of silver were prepared by glancing angle deposition technique (GLAD) described elsewhere.13 Briefly, the Si substrate was kept in a vacuum chamber at an angle called glancing angle to the incident metal plume. The various parameters of the coated film, such as thickness, porosity, topography were controlled by suitably adjusting the temperature, pressure, tilt angle, rotation speed etc. It has already been reported that Ag n-STFs having about 300 nm in height and 30% porosity over silicon substrates possess the highest SERS enhancement.4,12,13 Therefore, the n-STFs with optimum performance were fabricated for the present study.Sensor chip development
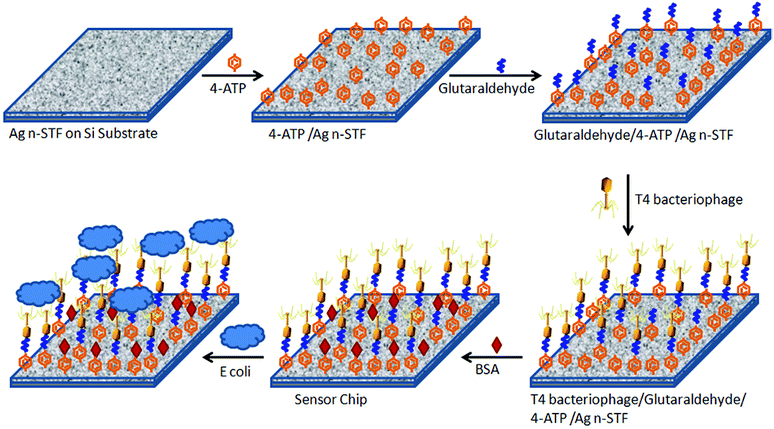
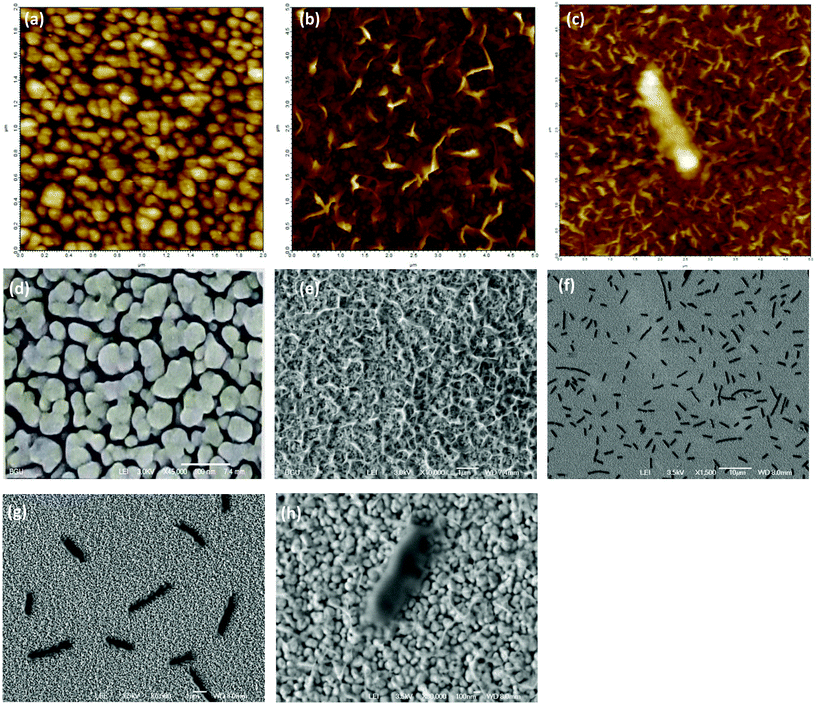
A small piece (about 5–7 mm2) of the fabricated n-STF was incubated overnight in 4-ATP solution in ethanol (1% w/w). As a result, a self-assembled monolayer of 4-ATP was spontaneously allowed to form over the Ag surface. The chip was then taken out of the 4-ATP solution and rigorously washed with ethanol and a continuous flow of water to remove any remnants. The chip was dried by blowing nitrogen gas. The chip was then incubated in aqueous solution of glutaraldehyde (1% v/v) for 1 hour to form a cross-linking layer. After taking the chip out of the glutaraldehyde solution, it was again washed with water to remove any unbound molecules and dried with a blow of nitrogen gas. The chips were then further incubated in T4 bacteriophage solution for 4 hours to form the specific receptor layer for the target E. coli B bacteria. Thereafter, the chip was incubated in BSA solution of 1 mg ml−1 concentration in 50 mM PBS buffer for 1 hour to block any remaining empty surface sites to prevent putative non-specific binding on the sensor surface. This step leads to an increase in specificity and hence the performance of the sensor, while reducing the effect of interference from other substances present in the sample which may add falsely to the signal. After BSA incubation, the chip was taken out, washed rigorously in water and PBS and blow dried with nitrogen. The chip was stored in a refrigerator at 4 °C prior to characterization. A schematic of the stepwise sensor chip fabrication is shown in Fig. 1.The atomic force microscope (AFM) and scanning electron microscope (SEM) images of the sensor chip before and after surface functionalization are shown in Fig. 2(a), (b), (d) and (e), respectively. In both the AFM (2(b)) and the SEM (2(e)) images, the leg like structures of the immobilized T4 bacteriophage are clearly visible at the sensor's surface after functionalization, while the n-STF surface is also visible in the background of the functionalized material. Fig. 2(e) and (f) show the SEM image of a number of E. coli attached to the sensor surface, while Fig. 2(g) and (h) show high resolution images of a single bacterium attached to the sensor surface. This clearly indicates that a single bacterium is attached to multiple phages immobilized on the sensor chip. On average, the sensor chips were 5–7 mm2 of size. Since the area of the focussed spot of the laser beam (in Fig. 3) was about 90 μm2, even smaller sensor chips up to dimensions of 100 μm2 are sufficient. However, greater dimensions were used due to the ease of handling. The images in Fig. 2(c) and (g) were taken after 30 minutes of interaction to see whether the bacteria became lysed with time or not.
Experimental setup
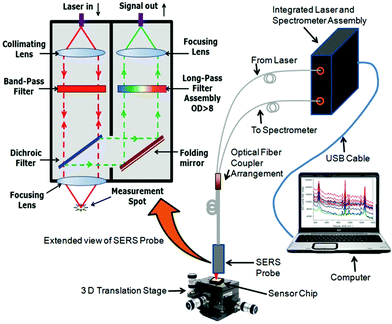
An illustration of the experimental setup is shown in Fig. 3. A fiber optic Raman spectrometer was used in these studies. It consists of both a laser and a spectrometer assembly housed in a single package. The light from a 785 nm laser was coupled to the proximal input end of a fiber optic coupler arrangement and the collected signal light from the far end of the coupler was fed to the spectrometer. Light from the far end output of the fiber optic SERS probe was focussed on the desired spot on the sensor chip with the help of a three dimensional (3D) translation stage. The captured enhancement is found to be maximum at the focus. The Stokes lines were captured from the same aperture and sent to the spectrometer via another optical fiber and suitable optics. The spectrometer was interfaced to a computer which further translated the captured signal into the SERS spectrum. An extended view of the SERS probe at the end of the optical fiber coupler is also shown in the same figure. As is evident, light from the laser is first collimated and then passed through a band pass filter to ensure the excitation at a single wavelength range and focussed on the sample by another lens. The enhanced Raman signals as well as the reflected light from the laser are captured by the same lens. The Raman signals are filtered from the reflected light by a dichroic filter and sent to the collecting assembly. The light from the dichroic filter is sent to a long pass filter assembly by a folding mirror. The long pass filter again filters the Rayleigh scattered light from the Raman scattered wavelengths. The Raman scattered light transmitted through the long pass filter assembly is then focussed on the input end of the collecting optical fiber by another lens. The operation principle of the sensor is that a change in the SERS enhancement of the sensor chip occurs when the bacteria are captured by the bacteriophage and thus brought intimately to the sensitive surface.As the SERS is a very short range phenomenon, we assessed only the SERS spectra from 4-ATP adsorbed on the surface of the n-STF. There is almost negligible change in the SERS spectra due to further binding of glutaraldehyde, bacteriophage and BSA. It has already been established that further binding does not affect much the Raman bands of the molecule nearest to the metal surface.4 Further binding of molecules may only result either in increase or decrease in the Raman enhancement. The change in enhancement can further be translated into the concentration of the binding analyte molecule. This is the working principle of such sensors. When the bacterium binds to the bacteriophage, it inserts its DNA into the bacterium which further replicates very quickly into the bacteria. After a certain amount of time (30 min), the bacterium is full of bacteriophages, that are about to burst. Certain sensors utilizing refractive index change operate on this principle so that the response could be recorded around the peak time, when the bacterial matrix is modified the maximum.22 However, it was also shown that if the interaction is carried out only up to the phage–E. coli association time, the mechanism can be used in SPR based detection methods.23 Such sensor surfaces could be regenerated for further use. In the present case, the bacterial association on the sensor surface was carried out for 10 minutes and the surface was regenerated for further use. The integration time of the spectrometer was 20 seconds.
The sample solutions of different concentrations ranging from 150 to 105 cfu ml−1 were prepared in PBS buffer from the stock solutions of E. coli B, P. aeruginosa and C. violaceum bacteria by making appropriate dilutions from stock solutions of 1.5 × 108, 3 × 107 and 1.5 × 109 cfu ml−1 concentrations, respectively. Similarly, the sample solutions of E. coli μX ranging from 340 to 105 cfu ml−1 and P. denitrificans ranging from 265 to 105 cfu ml−1 were prepared from the stock solutions of 3.4 × 108 and 5.3 × 108 cfu ml−1 concentrations respectively. The sensor chips were incubated in the sample solutions for 10 minutes and then taken out and washed with PBS buffer twice. The remaining moisture/liquid was removed by lightly and carefully blowing air. The excess liquid was removed to avoid any lensing effects due to the droplet on the sensor chip. The SERS spectra were then recorded from at least three places on the same sensor chip. After the recording of the spectra, the sensor chip was washed in running NaOH aqueous solution (20 mM) three minutes. This washing helps remove the attached bacteria from the surface and hence regenerate the sensor surface.23 The sensor surface was then rinsed twice with PBS to remove the remnants of the NaOH solution. The chip was blow dried again with air and incubated in other sample solution. This procedure was repeated for all the sample solutions at different concentrations of all kinds of bacteria used in the present study. The SERS spectra recorded from the three spots at the chip were averaged.
Results and discussion
The recorded SERS spectra for different concentrations of E. coli B are shown in Fig. 4. It is observed that there is a change in the SERS enhancements at different concentrations but no trend of variation can be predicted. However, when all the SERS spectra are referenced to the zero level, a clear trend in the variation of the enhancement with increase in the bacteria concentration is observed. As can be seen in the inset, where we have plotted the Raman enhancement of the 1077 cm−1 band for different concentrations of E. coli B, the enhancement increases with an increase in the bacterial concentration and becomes almost saturated. Similar SERS spectra were recorded for the varying concentrations of E. coli μX, P. denitrificans, P. aeruginosa and C. violaceum bacteria. The SERS spectra for CV026 have been plotted in Fig. 5. It is easily seen that almost no change in SERS enhancement was observed, even from the inset, which was similar to that for E. coli B, where we have plotted the Raman band at 1077 cm−1 for different bacterial concentrations.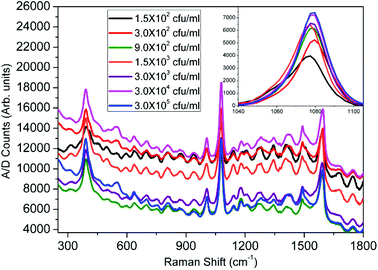 | ||
| Fig. 4 SERS spectra for varying concentrations of E. coli B. The inset shows the variation of Raman enhancement with concentration at 1077 cm−1. | ||
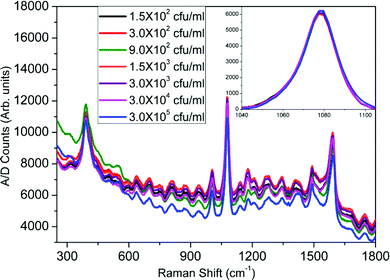 | ||
| Fig. 5 SERS spectra for varying concentrations of C. violaceum (CV026). The inset shows the variation of Raman enhancement with concentration at 1077 cm−1. | ||
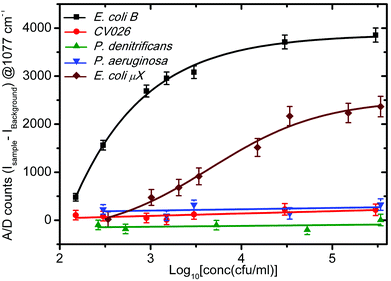
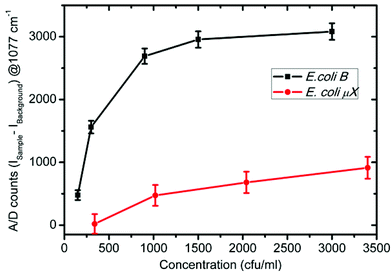
For a more quantitative prediction of the response of the sensor, we plotted the Raman enhancement versus concentration at 1077 cm−1 for all kinds of bacteria in Fig. 6. The Raman peak at 1077 cm−1 was selected because of the maximum enhancement at this band, which corresponds to the largest sensitivity and dynamic range of the sensor. The concentration at the x-axis is logarithmic due to the very large dynamic range of the sensor. The background Raman enhancement from the bare sensor chip was subtracted from the SERS signal of samples over the sensor to avoid any unavoidable deviations due to fluctuations in the background. Hence, the Y-axis represents the A/D counts from the sample minus that from the sensor (ISample − IBackground) at 1077 cm−1. Throughout the manuscript we designate the processed signal as the differential Raman enhancement. The symbols represent the differential Raman enhancement at different concentrations of different bacteria extracted from the respective SERS spectra, while the lines through them are the best curve fits. It is clear that with an increase in the concentration of both types of E. coli, the differential Raman enhancement first increases and then becomes nearly constant. However, for C. violaceum, P. denitrificans and P. aeruginosa there is almost no change in the differential Raman enhancement with increase in the concentrations. This confirms the specificity of the sensor for E. coli B detection. The successive addition of bacteria on the sensor surface contributes a little to the Raman enhancement, which leads to an increase in the signal. The region of saturation of the sensor response is less sensitive and hence puts a higher limit on the range of bacterial concentration, which can be detected by the sensor. This is due to the small size of the focussed laser beam, which limits the number of observed bacteria binding to the active region (exposed to the laser) of the sensor surface. Furthermore, it was observed that the sensitivity of the sensor is greater for E. coli B than that for E. coli μX. It may be due to a lower affinity of E. coli μX strain to T4 bacteriophage than E. coli B. Thus this possible lower affinity of E. coli μX may lead to reduced binding at the surface and hence to lower sensitivity. However, due to the reduced sensitivity, while the sensing spot remains the same size (= the spot size of the laser beam, 90 μm2) the dynamic range of the sensor becomes larger for E. coli μX. Furthermore, this sensor can detect whether an unknown sample contains any E. coli or not in the first instance, but an additional set of measurements are required to distinguish between different E. coli strains. To estimate the exact cell count and the E. coli strain, one should make further dilutions (at least one) of the unknown sample to see the matching of the response curve to that of one of the two E. coli bacterial strains. This provides an accurate measure of whether the sample solution contains E. coli or not and at what concentrations. We plotted the response of the sensor for very low bacterial concentrations varying from 150 to 3000 cfu ml−1 in Fig. 7. The error bars in Fig. 6 and 7 were calculated by taking into consideration the least count of the pipettes, and the noise level of the SERS measurement. It is quite clear from the plot (Fig. 7) that E. coli B concentrations down to 150 cfu ml−1 (∼1 bacterium per 10 μl) and E. coli μX concentrations down to 340 cfu ml−1 can be detected with the present sensor. The sample volumes in our experiments were kept at approximately 10 microliters, which means that such a volume when taken from a well-mixed sample of 150 cfu ml−1 will nearly always have at least a single bacterium in it. In this way, we can state that the sensor is capable to detect bacterial concentrations down to a single cell level in 10 μl volume of the sample. The Raman spectra for the sample solution concentrations below 150 cfu ml−1 were not analyzed to avoid any inconsistencies in the SERS signals, which may occur due to missing of even a single bacterium sometimes in the sample volume on the sensor surface.
Since, in general, the interaction between a T4 phage and E. coli results in the lysis of the bacteria, it might be possible to affect the signal of the sensor, if not properly monitored in time. To learn the fate of the bacteria on the sensor surface and understand the interaction with respect to time, we fabricated the sensor surface on glass micro-slides using a protocol similar to Tripathi et al.22 Briefly, the glass slides were treated in piranha solution (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, H2SO4
1, H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2) for five minutes, then rigorously washed with running DI water and finally blow dried under a nitrogen stream. Furthermore, a thin aminosilane layer was created over the glass slides by incubating them in a 1% (v/v) solution of trimethoxy aminosilane in (10
H2O2) for five minutes, then rigorously washed with running DI water and finally blow dried under a nitrogen stream. Furthermore, a thin aminosilane layer was created over the glass slides by incubating them in a 1% (v/v) solution of trimethoxy aminosilane in (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
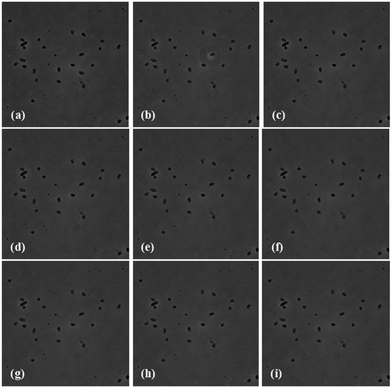
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) C2H5OH + CH3COOH solution. The acetic acid prevents the formation of multilayers of aminosilane.42 The glass slides were further incubated in 1% (v/v) aqueous solution of glutaraldehyde and then in the bacteriophage solution, as described earlier in the “Sensor chip development” section. The E. coli B suspension in PBS was poured on the micro-slide chip for 10 minutes and then washed twice with PBS to remove any uncaptured E. coli. Furthermore the sensor surface of the chip was immersed in PBS with the help of a 70 micron spacer and a cover slide. A schematic of the microscopic slide arrangement, which was used for time resolved phase contrast microscopy, is shown in Fig. 8. The captured images from the phase contrast microscope at different time intervals, over a period of 1 hour are shown in Fig. 9. It is quite evident that there is no change in bacterial count over time, indicating that E. coli cells are not lysed by the bacteriophages. Had the E. coli cells been lysed, the number of bacteria observed in the phase contrast microscope would have decreased. This result was supported by various reports already present in the literature.43–46 The reason behind this kind of observation is attributed to the fact that bacteria are lysed by the bacteriophage only in the growth culture media and when they are free floating. In the present case, since the T4 bacteriophages are immobilized and the bacteria are in PBS, a non-growth medium, no lysis occurs. It means that the results should not get affected with respect to time. Absorption of the phages to the surface may change the plasticity and the movement of the phage and prevent phage DNA injection. In order to inject DNA/RNA the phages need to exhibit conformational changes and contraction.43,44 Bacteriophages with contractile tails epitomize the concepts of “virus” and “phage” for many because the tails of these phages undergo a large conformational change – resembling the action of a syringe – upon the attachment to the host cell.
4) C2H5OH + CH3COOH solution. The acetic acid prevents the formation of multilayers of aminosilane.42 The glass slides were further incubated in 1% (v/v) aqueous solution of glutaraldehyde and then in the bacteriophage solution, as described earlier in the “Sensor chip development” section. The E. coli B suspension in PBS was poured on the micro-slide chip for 10 minutes and then washed twice with PBS to remove any uncaptured E. coli. Furthermore the sensor surface of the chip was immersed in PBS with the help of a 70 micron spacer and a cover slide. A schematic of the microscopic slide arrangement, which was used for time resolved phase contrast microscopy, is shown in Fig. 8. The captured images from the phase contrast microscope at different time intervals, over a period of 1 hour are shown in Fig. 9. It is quite evident that there is no change in bacterial count over time, indicating that E. coli cells are not lysed by the bacteriophages. Had the E. coli cells been lysed, the number of bacteria observed in the phase contrast microscope would have decreased. This result was supported by various reports already present in the literature.43–46 The reason behind this kind of observation is attributed to the fact that bacteria are lysed by the bacteriophage only in the growth culture media and when they are free floating. In the present case, since the T4 bacteriophages are immobilized and the bacteria are in PBS, a non-growth medium, no lysis occurs. It means that the results should not get affected with respect to time. Absorption of the phages to the surface may change the plasticity and the movement of the phage and prevent phage DNA injection. In order to inject DNA/RNA the phages need to exhibit conformational changes and contraction.43,44 Bacteriophages with contractile tails epitomize the concepts of “virus” and “phage” for many because the tails of these phages undergo a large conformational change – resembling the action of a syringe – upon the attachment to the host cell.
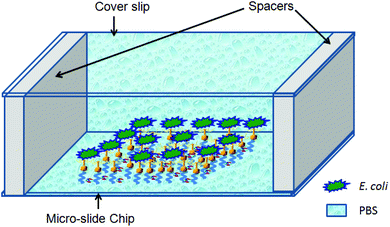 | ||
| Fig. 8 Schematic of the micro-slide chip for phase contrast microscopy (components are not to scale). | ||
Since the spot of the laser beam was about 90 microns, multiple micro-sized spots of the same sensor chip can be used for the detection of multiple samples. The nSTFs are very cost effective, as large, uniform chips can easily be fabricated. Additionally, the sensor chip is reusable, which makes it more cost effective. Other than that, the integration of this sensor chip with an optical fiber makes it capable for remote sensing and field applications. Furthermore, the sensor shown herein is faster and more sensitive than many commercially available E. coli detection kits.14
Conclusions
We have fabricated a nanobiosensor chip for the specific and quantitative detection of two different strains of E. coli. The sensor utilizes SERS based detection over optimized nSTFs of Ag over Si. Control experiments with C. violaceum, P. denitrificans and P. aeruginosa confirmed the lack of non-specificity of the sensor. The time resolved phase contrast microscopy confirmed that no lysis of bacteria occurs on the sensor chip, which further supports that our measurements are time independent. The sensor could detect E. coli B concentrations down to 1.5 × 102 cfu ml−1, which is an order of magnitude lower than a recently reported SERS sensor.28 The sensor utilizes low volumes (10 μl) of the sample solution. In addition, it was shown to be capable of detecting E. coli B concentrations down to a single bacterium. One should note that phages can be stored for a long time on a surface, making the system amenable to long-term storage as a product.Acknowledgements
This research is conducted by NTU-HUJ-BGU Nanomaterials for Energy and Water Management Programme under the Campus for Research Excellence and Technological Enterprise (CREATE), that is supported by the National Research Foundation, Prime Minister's Office, Singapore. The support by the Graduate school ‘BuildMoNa’ (University Leipzig, Germany) funded within the German Excellence Initiative of the Deutsche Forschungsgemeinschaft (DFG) is also appreciated. Sachin K Srivastava thanks the Council of Higher Education of the Government of the State of Israel for PBC post-doctoral fellowship.References
- K. E. Shafer-Peltier, C. L. Haynes, M. R. Glucksberg and R. P. Van Duyne, J. Am. Chem. Soc., 2002, 125, 588–593 CrossRef PubMed.
- J. Chen, J. Jiang, X. Gao, G. Liu, G. Shen and R. Yu, Chem. – Eur. J., 2008, 14, 8374–8382 CrossRef CAS PubMed.
- H. T. Ngo, H.-N. Wang, A. M. Fales and T. Vo-Dinh, Anal. Chem., 2013, 85, 6378–6383 CrossRef CAS PubMed.
- S. K. Srivastava, A. Shalabney, I. Khalaila, C. Grüner, B. Rauschenbach and I. Abdulhalim, Small, 2014, 10, 3579–3587 CrossRef CAS.
- S. A. Kalele, A. A. Kundu, S. W. Gosavi, D. N. Deobagkar, D. D. Deobagkar and S. K. Kulkarni, Small, 2006, 2, 335–338 CrossRef CAS PubMed.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- P. L. Stiles, J. A. Dieringer, N. C. Shah and R. P. Van Duyne, Annu. Rev. Anal. Chem., 2008, 1, 601–626 CrossRef CAS PubMed.
- J. B. Jackson, S. L. Westcott, L. R. Hirsch, J. L. West and N. J. Halas, Appl. Phys. Lett., 2003, 82, 257–259 CrossRef CAS PubMed.
- C. J. Orendorff, L. Gearheart, N. R. Jana and C. J. Murphy, Phys. Chem. Chem. Phys., 2006, 8, 165–170 RSC.
- A. Tao, F. Kim, C. Hess, J. Goldberger, R. He, Y. Sun, Y. Xia and P. Yang, Nano Lett., 2003, 3, 1229–1233 CrossRef CAS.
- T. Wang, X. Hu and S. Dong, J. Phys. Chem. B, 2006, 110, 16930–16936 CrossRef CAS PubMed.
- A. Shalabney, C. Khare, J. Bauer, B. Rauschenbach and I. Abdulhalim, J. Nanophotonics, 2012, 6, 061605 CrossRef PubMed.
- I. Abdulhalim, Small, 2014, 10, 3499–3514 CrossRef CAS PubMed.
- MicroSEQ® E. coli O157:H7 Detection Kit, http://tools.lifetechnologies.com/content/sfs/brochures/CO13764.pdf, Accessed 10–09–2014, 2014.
- CNN, E. coli Outbreaks Fast Facts, http://edition.cnn.com/2013/06/28/health/e-coli-outbreaks-fast-facts/, Accessed 10–09–2014, 2014.
- I. Abdulhalim, A. Karabchevsky, C. Patzig, B. Rauschenbach, B. Fuhrmann, E. Eltzov, R. Marks, J. Xu, F. Zhang and A. Lakhtakia, Appl. Phys. Lett., 2009, 94, 063106 CrossRef PubMed.
- D. D. Deobagkar, V. Limaye, S. Sinha and R. D. S. Yadava, Sens. Actuators, B, 2005, 104, 85–89 CrossRef CAS PubMed.
- K. A. Fode-Vaughan, J. S. Maki, J. A. Benson and M. L. P. Collins, Lett. Appl. Microbiol., 2003, 37, 239–243 CrossRef CAS.
- P. M. Fratamico, T. P. Strobaugh, M. B. Medina and A. G. Gehring, Biotechnol. Tech., 1998, 12, 571–576 CrossRef CAS.
- J. Gau Jr., E. H. Lan, B. Dunn, C.-M. Ho and J. C. S. Woo, Biosens. Bioelectron., 2001, 16, 745–755 CrossRef.
- B. Guven, N. Basaran-Akgul, E. Temur, U. Tamer and I. H. Boyac, Analyst, 2011, 136, 740–748 RSC.
- S. M. Tripathi, W. J. Bock, P. Mikulic, R. Chinnappan, A. Ng, M. Tolba and M. Zourob, Biosens. Bioelectron., 2012, 35, 308–312 CrossRef CAS PubMed.
- S. K. Arya, A. Singh, R. Naidoo, P. Wu, M. T. McDermott and S. Evoy, Analyst, 2011, 136, 486–492 RSC.
- A. Karabchevsky, C. Khare, B. Rauschenbach and I. Abdulhalim, J. Nanophotonics, 2012, 6, 061508 CrossRef PubMed.
- K. Abu-Rabeah, A. Ashkenazi, D. Atias, L. Amir and R. S. Marks, Biosens. Bioelectron., 2009, 24, 3461–3466 CrossRef CAS PubMed.
- A. Golberg, G. Linshiz, I. Kravets, N. Stawski, N. J. Hillson, M. L. Yarmush, R. S. Marks and T. Konry, PLoS One, 2014, 9, e86341 Search PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- J. Chen, X. Wu, Y.-w. Huang and Y. Zhao, Sens. Actuators, B, 2014, 191, 485–490 CrossRef CAS PubMed.
- Y. C. Cao, R. Jin and C. A. Mirkin, Science, 2002, 297, 1536–1540 CrossRef CAS PubMed.
- Q. Pan, X.-L. Zhang, H.-Y. Wu, P.-W. He, F. Wang, M.-S. Zhang, J.-M. Hu, B. Xia and J. Wu, Antimicrob. Agents Chemother., 2005, 49, 4052–4060 CrossRef CAS PubMed.
- A. Sengupta, M. Mujacic and E. J. Davis, Anal. Bioanal. Chem., 2006, 386, 1379–1386 CrossRef CAS PubMed.
- A. Singh, N. Glass, M. Tolba, L. Brovko, M. Griffiths and S. Evoy, Biosens. Bioelectron., 2009, 24, 3645–3651 CrossRef CAS PubMed.
- S. Balasubramanian, I. B. Sorokulova, V. J. Vodyanoy and A. L. Simonian, Biosens. Bioelectron., 2007, 22, 948–955 CrossRef CAS PubMed.
- B. Van Dorst, J. Mehta, K. Bekaert, E. Rouah-Martin, W. De Coen, P. Dubruel, R. Blust and J. Robbens, Biosens. Bioelectron., 2010, 26, 1178–1194 CrossRef CAS PubMed.
- T. K. Van Dyk, W. R. Majarian, K. B. Konstantinov, R. M. Young, P. S. Dhurjati and R. A. Larossa, Appl. Environ. Microbiol., 1994, 60, 1414–1420 CAS.
- Y. Dessaux, C. Elmerich and D. Faure, Rev. Med. Int., 2004, 25, 659–662 CrossRef CAS PubMed.
- K. H. McClean, M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart and P. Williams, Microbiology, 1997, 143, 3703–3711 CrossRef CAS PubMed.
- S. C. Baker, S. J. Ferguson, B. Ludwig, M. D. Page, O.-M. H. Richter and R. J. M. van Spanning, Microbiol. Mol. Biol. Rev., 1998, 62, 1046–1078 CAS.
- L. Bergaust, Y. Mao, L. R. Bakken and Å. Frostegård, Appl. Environ. Microbiol., 2010, 76, 6387–6396 CrossRef CAS PubMed.
- M. M. Moritz, H.-C. Flemming and J. Wingender, Int. J. Hyg. Environ. Health, 2010, 213, 190–197 CrossRef CAS PubMed.
- S.-E. Hsieh, H.-H. Lo, S.-T. Chen, M.-C. Lee and Y.-H. Tseng, Appl. Environ. Microbiol., 2011, 77, 756–761 CrossRef CAS PubMed.
- S. K. Srivastava, V. Arora, S. Sapra and B. D. Gupta, Plasmonics, 2012, 7, 261–268 CrossRef CAS.
- P. Leiman and M. Shneider, in Viral Molecular Machines, ed. M. G. Rossmann and V. B. Rao, Springer, US, 2012, vol. 726, pp. 93–114 Search PubMed.
- M. G. Rossmann, V. V. Mesyanzhinov, F. Arisaka and P. G. Leiman, Curr. Opin. Struct. Biol., 2004, 14, 171–180 CrossRef CAS PubMed.
- M. Los, P. Golec, J. Los, A. Weglewska-Jurkiewicz, A. Czyz, A. Wegrzyn, G. Wegrzyn and P. Neubauer, BMC Biotechnol., 2007, 7, 13 CrossRef PubMed.
- H. Hadas, M. Einav, I. Fishov and A. Zaritsky, Microbiology, 1997, 143, 179–185 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |