Bicontinuous cubic phase nanoparticle lipid chemistry affects toxicity in cultured cells†
Tracey M.
Hinton‡
a,
Felix
Grusche‡
a,
Durga
Acharya
b,
Ravi
Shukla
c,
Vipul
Bansal
c,
Lynne J.
Waddington
b,
Paul
Monaghan
a and
Benjamin W.
Muir
*b
aCSIRO Animal, Food and Health Sciences, Australian Animal Health Laboratory, 5 Portarlington Rd, East Geelong, VIC 3219, Australia
bCSIRO Materials Science and Engineering, Bayview Avenue, Clayton, VIC 3168, Australia. E-mail: ben.muir@csiro.au
cNanoBiotechnology Research Lab, School of Applied Sciences, RMIT University, Melbourne, VIC 3001, Australia
First published on 18th October 2013
Abstract
Gaining an increased understanding of the toxicity of new lipid nanoparticle formulations such as the class of cubic and hexagonal phase forming nanomaterials called cubosomes™ and hexosomes™ is crucial for their development as therapeutic agents. Surprisingly, the literature on the in vitro and in vivo toxicity of cubic and hexagonal phase forming lipid nanoparticles is negligible, despite a rapidly growing number of publications on their potential use in various therapeutic applications. In this work we have developed methods to study the in vitro cytotoxicity of two chemically distinct cubic phase nanoparticle dispersions using the lipids glycerol monooleate and phytantriol respectively. We have found that the toxicity of phytantriol cubosomes is considerably greater than that of glycerol monooleate cubosomes. The increased toxicity of phytantriol appears to result from its greater ability to disrupt the cellular membrane (haemolytic activity) and oxidative stress. This finding has significant impact and can provide useful guidelines for those conducting further research on the use of cubic phase forming lipids for therapeutic and diagnostic applications both in vitro and in vivo.
Introduction
The self assembly of amphiphilic molecules to produce lyotropic liquid crystal phases is a broad area of research.1,2,4–7 The common phases seen when using lyotropic liquid crystal forming materials include lamellar phase comprised of bilayer sheets (periodicity in one dimension),8,9 the hexagonal phase which consists of infinitely long hexagonally packed rods (periodicity in two dimension), and cubic phases consisting of either discrete micelles arranged into a cubic lattice (discontinuous cubic structure), or interwoven bicontinuous hydrophilic and hydrophobic domains with long range periodicity in three dimensions.6 The bicontinuous cubic phases have a number of closely related structures, where the underlying crystal lattice can be described by the gyroid (G), diamond (D) and primitive (P) minimal surfaces, which correspond to the Ia3d (G), Pn3m (D) and Im3m (P) crystallographic space groups, respectively. Such self-assembled scaffolds with three dimensional structure exhibit extensive porosity and remarkably high surface areas. Of specific interest is the use of lipids to generate these complex structures.7,10 Dispersions of hexagonal phase nanoparticles are commonly called hexosomes™ and cubic phase nanoparticles are referred to as cubosomes™.6Lipids such as glycerol monooleate (GMO, also known as monoolein) and phytantriol (Phy) form inverse bicontinuous cubic phases (Q2) when exposed to water, with Ia3d crystal lattice structure at low levels of hydration and Pn3m crystal lattice structures at high levels of hydration.11 These optically isotropic and highly viscous gel-like materials exhibit good temperature stability and can solubilize both hydrophobic and hydrophilic compounds into their structure.6 The incorporation of additives to such materials may result in the formation of other phases such as the inverse hexagonal phase (H2) due to an alteration in the spontaneous curvature of the lyotropic liquid crystal assemblies. These Q2 and H2 phases in excess water can be colloidally dispersed into stable nanoparticles after application of mechanical or ultrasonic energy in presence of stabilizer such as the Pluronic® F127 block copolymer.12
Glycerol monooleate (GMO) and phytantriol (Phy) are commonly used in bulk form in food products11 and cosmetic products such as make up and shampoo. Glycerol monooleate contains an ester group that is susceptible to hydrolysis and therefore biodegradation in vivo. These cubosome nanomaterials are being extensively studied as potential therapeutic agents for drug/gene delivery and medical imaging applications.3–5,7,10,13–21 Despite a significant amount of effort investigating the use of such materials for therapeutic applications,22 minimal research has been undertaken to investigate potential toxic effects of nanoparticle formulations made from lyotropic liquid crystal nanoparticles.23–25 The toxicity of nanoparticles is known to be influenced by size, proliferation and embryonic origin of the cells which are chosen for testing.26–31 The toxicity of a variety of lipid based nanoparticles have been well tested32 but few reports of the toxicity of lyotropic liquid crystal phase forming lipid nanoparticles exist. A few studies showing the possible membrane disruption ability of cubic phase lipid nanoparticles23 and toxic effects in vitro24,25 have been reported. In this work we present a study of the toxicity, in cultured and primary cells, of two cubic phase forming lipids, namely GMO and Phy cubosomes. As has been demonstrated before, we note a significant difference in the cytotoxicity profile of GMO vs. Phy cubosomes.33 We note differences in the subcellular distribution of GMO and Phy cubosomes and find that Phy cubosomes result in a disruption of cell membrane integrity and an inhibition of cell membrane remodeling via macropinocytosis.
Experimental
Materials
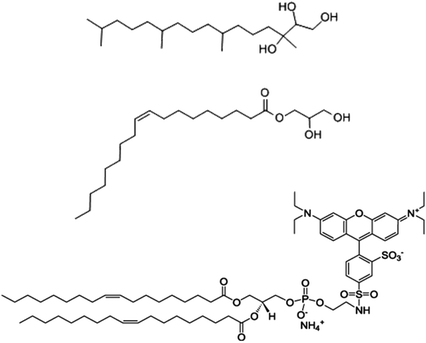
Phytantriol (Phy) (98.0%) was obtained from DSM Nutritional Products. Glycerol monooleate (GMO) was obtained from Nu-Check-Prep Inc. (99.0%), octadecyl rhodamine B chloride (R18) was obtained from Invitrogen and Pluronic® F127 (poly(ethylene oxide)-poly(propyleneoxide)-poly(ethylene oxide)) was purchased from Sigma and used as received (Fig. 1). All chemicals were used without purification.Synthesis of cubosome™ nanoparticles
Two sets of each cubosome formulation was prepared, cubosomes not containing dye were used for the lucifer yellow and macropinocytosis assays. All other experiments were conducted using cubosomes containing dye. For the R-18 dye containing cubosomes, in a glass vial 15 μg of Octadecyl Rhodamine B chloride (R18) dye was taken as a solution in chloroform and the solvent was removed completely under a stream of nitrogen for about 1 h at 40 °C. To this 50 mg of Phy or GMO was added and mixed well at 40 °C. Two ml of water containing 9 mg of F-127 was added to the vial containing the mixture of lipid and R18 lipid dye. The Pluronic polymer F-127 acts as a polymeric steric stabiliser via hydrophobic interactions of the propyleneoxide units with the lipids in the cubosomes. The mixture was heated to 70 °C and dispersed via ultrasonication (Misonix Sonicator S-4000 attached with a Misonix microtip 418) for 3 min in pulse mode (2 s sonic pulses interrupted by 3 s breaks) at 50% of maximum amplitude of the equipment. This resulted in the formation of pink solutions which were stored at 25 °C prior to further experimentation.Particle size measurements
Particle size measurements were performed in phosphate buffered saline (PBS) (100 mM) using a Zetasizer-Nano instrument (Malvern, UK). Measurements were conducted at 25 °C, using automated settings in standard disposable cuvettes. Ten measurements were made per sample.Cryo-TEM measurements
Samples for cryo-TEM studies were examined using a Gatan 626 cryoholder (Gatan, Pleasanton, CA, USA) and Tecnai 12 Transmission Electron Microscope (FEI, Eindhoven, The Netherlands) at an operating voltage of 120 kV. Images were recorded using FEI Eagle 4k × 4k CCD camera, at magnifications ranging from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× to 50
000× to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×. Samples were prepared on 200-mesh copper grids coated with perforated carbon film (Lacey carbon film: ProSciTech, Qld, Australia) using a method which has been reported previously.12
000×. Samples were prepared on 200-mesh copper grids coated with perforated carbon film (Lacey carbon film: ProSciTech, Qld, Australia) using a method which has been reported previously.12
Cell lines
Chinese Hamster Ovary cells (CHO: ATCC CCL-061) were grown in MEMα modification, and human alveolar basal epithelial cells (A549; ATCC CCL-185) were grown in DMEM. Both base media were supplemented with 10% foetal bovine serum, 2 mM glutamine, 10 mM Hepes, 1.5 g l−1 sodium bicarbonate, 0.01% penicillin and 0.01% streptomycin. Cells were grown at 37 °C with 5% CO2 and subcultured twice weekly.Toxicity assay
CHO and A549 cells were seeded at 1 × 104 cells per well in 96-well tissue culture plates and grown overnight at 37 °C with 5% CO2.The lipid formulations at concentrations varying from 100 to 3.12 μg ml−1 were added to 3 wells in the 96 well culture plates for each sample and incubated for 72 h at 37 °C in 200 μl standard media. Toxicity was measured using the Alamar Blue reagent (Invitrogen USA) according to manufacturer's instructions. Briefly, media was removed, cells were washed once with PBS and replaced with 100 μl of standard media containing 10% Alamar Blue reagent, cells were then incubated for 4 h at 37 °C with 5% CO2. The assay was read on an EL808 Absorbance microplate reader (BIOTEK, USA) at 540 nm and 620 nm. Cell viability was determined by subtracting the 620 nm measurement from the 540 nm measurement. Obtained data were analysed in Microsoft Excel. Results are presented as a percentage of untreated cells and the presented data are representative of three separate experiments in triplicate ± SEM.
Imaging of cells for cubosome uptake, membrane disruption, and macropinocytosis inhibition assays
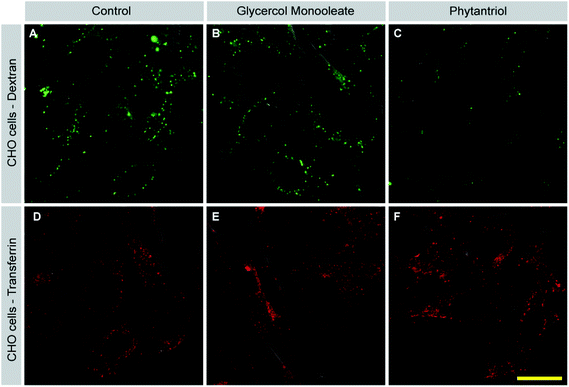
CHO and A549 cells were grown to a density of approximately 70% coverage on 13 mm round glass coverslips (Menzel, Germany) in 24 well plates (Nunc, USA). For cubosome uptake experiments, 20 μg ml−1 Phy- or GMO-containing, fluorescently labelled cubosomes were added to cells for 24 h. Cells were then processed for confocal imaging. For membrane disruption assays complete culture medium was replaced with OPTIMEM (Invitrogen, USA). Lucifer yellow (Sigma, USA) was added at a final concentration of 100 μg ml−1 and cells were cultured in the presence or absence of 20 μg ml−1 Phy- or GMO-containing cubosomes for one hour. As a positive control, cell membranes were mechanically disrupted by scraping a pipette tip across the coverslip prior to incubation in the presence of LY for one hour. For macropinocytosis disruption assays, complete culture medium was replaced with OPTIMEM and cells were incubated in the presence or absence of 20 μg ml−1 Phy- or GMO-containing cubosomes for 45 min. FITC-Dextran (Sigma, USA) was then added at a final concentration of 50 nM for 15 min to assay macropinocytic activity of cells. To measure clathrin-mediated endocytosis, Alexa-Fluor 568-labelled human Transferrin (Invitrogen, USA) was added to cells at a final concentration of 1 μg ml−1 for 10 min.To process cells for confocal microscopy, cells were washed in PBS and fixed in 4% paraformaldehyde (Sigma, USA) in PBS for one hour. Coverslips with cells were mounted onto slides in Vectashield (Vector Laboratories, USA). Images were acquired on a Leica SP5 confocal microscope (Leica Microsystems, Germany). All samples were processed in parallel and laser and gain settings for cubosomes, LY, FITC-Dextran and 568-Transferrin were kept constant between samples. All confocal images are single slices.
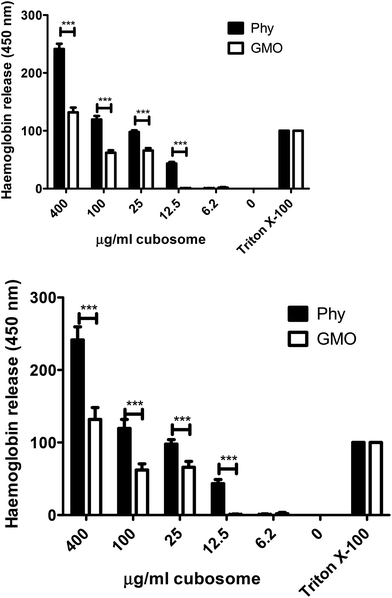
Haemolysis assay
Mouse blood in EDTA was obtained from C57/Blk6 mice from the AAHL small animal facility according to animal ethics committee (AEC) approval. The blood was washed in PBS three times and resuspended at a concentration of 7.5 × 107 cells per ml. Aliquots of 100 μl were added to 100 μl of PBS containing the serial dilution of cubosomes in a 96-well microtiter plate and incubated for 1 h at 37 °C with constant shaking. After removal of the unlysed erythrocytes by centrifugation (1000g, 5 min), 100 μl of the supernatant were transferred to a new microtiter plate and haemoglobin absorption was determined at 450 nm (background correction at 750 nm) on an EL808 Absorbance microplate reader (BIOTEK, USA). 100% lysis was determined by adding 5 μl of a 0.1% Triton X-100 solution prior to centrifugation. Results are presented as percentage haemoglobin release compared to the PBS control.IFN-α and iNOS upregulation in primary mouse splenocytes
Spleens were removed from C57/Black 6 mice from the AAHL small animal facility according to AEC approval and purified splenocytes were prepared by sieving the spleens through a 70 μm cell strainer (BD Biosciences, USA) and layering onto lymphoprep (Vital Diagnostics, Australia). The cells were then centrifuged at 1200g for 20 min. The lymphocyte rich layer was removed and washed in PBS. RNA was harvested using the Trizol method.34Reverse transcription and quantitative real-time PCR
After RNA was harvested using the Trizol method,34 one microgram was treated with DNase (Promega, Madison, WI) according to the manufacturer's instructions. Quantitative real-time PCR (QRT-PCR) experiments were conducted using power SYBR green RNA to CT kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions to measure cytokine expression levels. Mouse Allalpha and GAPDH sequences were from ref. 35. Mouse iNOS 2 sequences were adapted from ref. 36, sequences of all primers are in Table 1. Primers were obtained from Geneworks (South Australia). All quantification data was normalised against mouse GAPDH. QRT-PCR was performed on a StepOnePlus Real Time-PCR System, 96 well plate RT-PCR instrument (Applied Biosystems) under the following conditions: 1 × cycle 50 °C for 30 min followed by 95 °C for 10 min, 40 × cycles 95 °C for 15 s followed by 60 °C for 1 min. The comparative threshold cycle (ΔΔCt) method was used to derive fold change gene expression. Data was normalised to mouse GAPDH.| Sequence | Primer sequence 5′–3′ | Sequence position |
|---|---|---|
| Mouse GAPDH F | ATCTTCTTGTGCAGTGCCAGC | 13–33 |
| Mouse GAPDH R | ACTCCACGACATACTCAGCACC | 315–336 |
| Mouse Allalpha F | SAWCYCTCCTAGACTCMTTCTGCA | 305–328 |
| Mouse IFNalpha Ra | TATDTCCTCACAGCCAGCAG | 418–437 |
| Mouse iNOS F | CCTCCAGTGTCTGGGAGCATCAC | 1561–1583 |
| Mouse iNOS R | CTCAGCTTCTCATTCTGCC | 1667–1685 |
Nitrate production assay
Nitric oxide (NO) production in supernatant was measured using a NO colorimetric assay kit (Roche cat no. 11756281001). The assay was conducted as per the manufacturer's instructions using a NO control with a standard curve plotted and samples were measured at a wavelength of 550 nm.Statistics
The difference between two groups was statistically analysed by one way repeated measures ANOVA, parametric, with Tukey post analysis. ***p < 0.001, *p < 0.05, N.S. not significant.RT-PCR gene expression analysis in response to GMO and PHY nanoparticles
To analyse expression of stress and apoptosis related genes in response to GMO and PHY, quantitative RT-PCR studies were performed. The cells were treated with nanoparticles for 24 h, RNA was isolated and cDNA was synthesized from 1 μg total RNA using random hexamer primers provided in the kit following manufacturer’ instructions.Quantitative RT-PCR was achieved by cDNA (100 ng per reaction) amplification and relative levels of target gene expression were measured on the 7500 Fast Real-time RT-PCR system (Applied Biosystems). For fluorescence measurement and assay preparation, FAM-490-based Taqman Gene Expression Assay Mix (Applied Biosystems) specific for each gene of interest and Taqman Universal Master Mix (Applied Biosystems) were used. Relative Quantification PCR analysis was performed using the ABI 7500 Software. Relative quantitation of target gene expression in A549 cells was done by using the comparative Ct method. The Ct value was defined as the cycle number at which the PCR amplification graph passed a threshold, which was auto-calculated by software for the measurements. The amount of targets and endogenous reference (β-actin) were determined from the standard curve. Target values were normalized to the endogenous reference, assuming that β-actin expression was identical in the different samples. The relative amount of target gene mRNA was calculated by using the formula: 2−ΔΔCt, where ΔΔCt = [Cttarget gene − Ctβ-actin] − [Ctβ-actin]. For β-actin, ΔΔCt equals zero and 20 equals one. For other genes, the value of 2−ΔΔCt indicates the fold change in gene expression relative to the β-actin controls.
Results and discussion
Nanoparticle characterisation
The structures of Phy, GMO and Rhodamine (R18) used in this work can be seen in Fig. 1. The R18 lipid was used to label the cubosomes as its hydrophobic chains can mix with the cubosome bulk lipid lipophilic domains to minimise release of the fluorophore. This made R18 an ideal candidate to be used as a fluorescent marker within the cubosomes while not adversely affecting their physicochemical properties such as liquid crystal phase, surface charge and morphology as shown in previous work.25 The size of the labelled cubosomes were measured by dynamic light scattering (DLS, intensity vs. size) (Fig. 2). Analysis with this technique showed that the two bulk lipid formulations produced nanoparticles (milky pink dispersions) that were remarkably similar in size. Specifically the GMO cubosomes produced nanoparticles with an average diameter of 190 nm and had a polydispersity index (PDI) of 0.14 while Phy cubosomes were measured to have an average diameter of 189 nm and PDI of 0.16. This finding is in agreement with use in previous studies where the nanoparticle sizes and polydispersity indices did not vary significantly between the two bulk lipids Phy and Myverol (the bulk of which is made up of GMO).12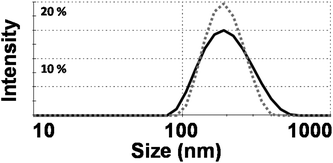 | ||
| Fig. 2 Dynamic light scattering (DLS) data of phytantriol (Phy) (dashed grey line) and glycerol monooleate (GMO) (solid black line) cubosomes (size vs. intensity). | ||
To further investigate the dispersed lyotropic liquid crystalline nanoparticles produced, cryo-TEM and confocal microscopy was used to image individual cubosomes. The results of this study can be seen in Fig. 3. The cryo-TEM images confirm the presence of discrete colloidal nanoparticles spanning a size range of between 60 nm and 300 nm which is typical of these systems and is consistent with the DLS traces observed. The Phy and GMO cubosomes show nanostructured particles displaying a periodic internal structure typical of cubic phase materials. The cryo TEM of the GMO cubosomes is typical of these nanoparticles. Of note is the fact that few attached vesicles are observed in Phy cubosomes while they are commonly observed in GMO cubosomes.
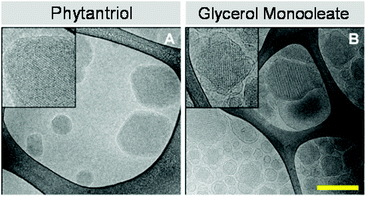 | ||
| Fig. 3 Cryo-TEM images of phytantriol (A) and glycerol monooleate (B) cubosomes. Insets on cryo-TEM images show periodic internal structure typical of cubic phase materials. Scale bar is 200 nm. | ||
Cytotoxicity of GMO and Phy cubosomes
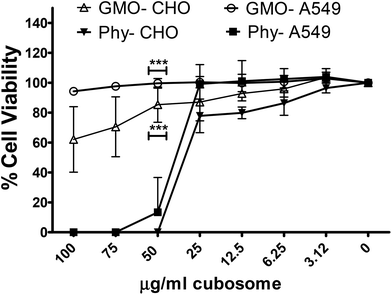
Previous cytotoxicity studies by us on phytantriol and Myverol based lyotropic mesophase nanoparticles demonstrated a higher toxicity of phytantriol containing nanoparticles compared to an equivalent concentration of Myverol nanoparticles in two different cell lines.12 To determine if this trend was observed between Phy and GMO cubosomes lipid formulations were produced containing equal amounts of either phytantriol or GMO and assayed for cytotoxicity in CHO and A549 cell lines. As expected GMO was non-toxic at a concentration of 100 μg ml−1 in the A549 cell line, whilst there was approximately 40% toxicity at 100 and 75 μg ml−1 in CHO-GFP cells. There was no toxicity in either cell line at 50 μg ml−1 based on an 80% viability threshold (Fig. 4).33 The phytantriol nanoparticle formulation was non-toxic at 25 μg ml−1 for both CHO cells and A549 cell lines (Fig. 4) however, the formulation was highly toxic at higher concentrations.Cellular uptake of GMO and Phy cubosomes
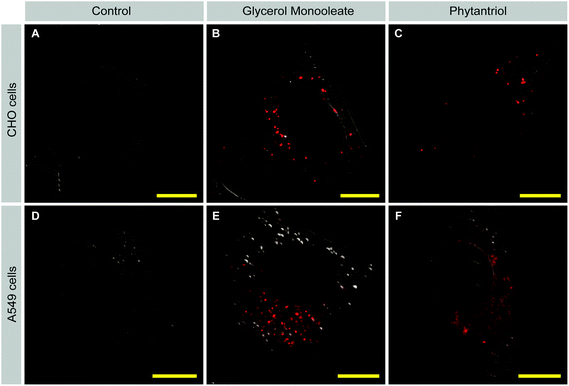
To investigate cellular uptake and subcellular distribution of cubosomes, fluorescently labelled cubosomes were added to CHO and A549 cells (Fig. 5). Following 24 hours of incubation, cells were washed and fixed. Samples were then mounted and imaged without further treatment to minimize potential artefacts due to permeabilizing and processing of cells. Differential interference contrast (DIC) was used to illuminate the structure of cells while the cubosomes were visualized based on their rhodamine-containing R18 lipid dye. Both GMO and Phy cubosomes were found inside CHO and A549 cells. In CHO cells, the fluorescent signal in both GMO and Phy cubosome-treated cells was visible as discrete punctate, suggesting a vesicular localisation (Fig. 5B and C). In addition, Phy cubosome treated but not GMO cubosome treated cells showed a diffuse cytoplasmic staining pattern with the percentage of cells containing diffuse lucifer yellow in CHO-WT was 27% whilst A549 cells were more affected with 52% containing the diffuse lucifer yellow signal. This percentage is not surprising as not all cells will take up equal amounts of cubosomes, therefore we do not expect all cells to show this result. In A549 cells, GMO cubosome treated cells showed a fluorescent signal that was punctate with some diffuse staining, while Phy cubosome treated cells showed a largely diffuse staining pattern (Fig. 5E and F). The punctate staining pattern of GMO treated cells suggests that GMO cubosomes were primarily localized within membrane-enclosed areas such as endosomes or lysosomes. On the contrary, the diffuse staining pattern that was seen in addition to the punctate staining pattern in Phy cubosome treated cells probably arose through two possible mechanisms: either some Phy cubosomes were able to freely move within the cytoplasm or the lipid material of the Phy cubosomes (namely phytantriol and R18 lipid dye) mixed with cellular membranes. Despite no cubosomes being visualised in the nucleus the lucifer yellow was. This is unlikely to be due to nuclear membrane disruption by the Phy cubosomes as lucifer yellow is able to enter the nucleus via the nuclear pores.Cell membrane disruption by Phy but not GMO cubosomes
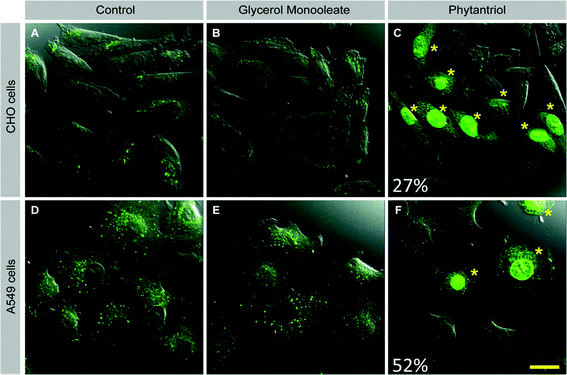
One possible reason for the increased toxicity of Phy-containing cubosomes as compared to GMO cubosomes is that Phy cubosomes show a higher degree of membrane disruption than GMO cubosomes. To test this hypothesis, we devised a membrane integrity assay based on exclusion of the fluorescent dye lucifer yellow, a low molecular weight dye (MW 457.2) that cannot pass intact cell membranes. Mechanical disruption of cell membranes by sliding a sharp object along a cell culture dish has been reported to result in dye uptake.37 We reasoned that if membrane disruption occurred with Phy cubosomes, then we might be able to visualize this through the presence of dye within the cells.CHO and A549 cells were therefore incubated in the presence of lucifer yellow with either Phy- or GMO cubosomes or in the absence of cubosomes for one hour. Cells were then fixed and analyzed by confocal microscopy. In both cell lines, control- and GMO-treated cells showed uptake of lucifer yellow only through endocytosis, visible as distinct green punctate throughout the cytoplasm (Fig. 6A, D and Fig. 6B, E, respectively). Strikingly, a proportion of Phy-treated cells showed strong, diffuse staining for lucifer yellow in the cytoplasm and nucleus (Fig. 6C and F). The diffuse staining pattern and presence of LY in the nucleus in Phy treated cells is distinct from the pattern seen for control- and GMO-treated cells and suggests that membrane disruption occurred, allowing diffusion of dye-containing extracellular fluid across the membrane of Phy treated cells. Importantly, the morphology of the LY positive cells was normal, indicating that membrane disruption was not a secondary effect of cell death/apoptosis.
To further assess the ability of GMO and Phy to disrupt cellular membranes a haemolysis assay was performed on serial dilutions of both cubosomes. In this assay, release of haemoglobin from red blood cells due to membrane disruption was measured. At all but the lowest concentration (6.2 μg ml−1) Phy resulted in a significantly higher release of haemoglobin than GMO (Fig. 7). This indicates that if an imaging agent or drug is delivered extremely efficiently to use minimal amounts of Phy cubosomes these may remain possible delivery agents.
Macropinocytosis inhibition by Phy but not GMO cubosomes
Our membrane disruption assay demonstrated that Phy led to membrane disruption at least in a subset of cells. In addition, we noticed that Phy treated cells with intact membranes contained fewer LY positive vesicles than control or GMO-treated cells. This observation indicated that cellular uptake mechanisms such as macropinocytosis are affected by Phy. To address this possibility, we assessed uptake of fluorescently labeled Dextran (FITC-Dextran), a specific marker for macropinocytosis, following treatment of cells with either GMO or Phy, or in untreated control cells (Fig. 8). Control and GMO cells showed comparable rates of Dextran uptake (Fig. 8A and B). Strikingly, Dextran uptake was strongly reduced in Phy-treated cells (Fig. 8C).Cells also internalize material through processes other than macropinocytosis such as clathrin-mediated endocytosis (CME) and caveolin-mediated endocytosis. To test whether Phy acted as a general inhibitor of endocytosis, we assessed inhibition of CME following cubosome treatment. After incubation with GMO or Phy, fluorescently labeled Transferrin (TF-568) was added to cells for 10 min. Transferrin is selectively taken up via CME. In contrast to Dextran uptake, Transferrin uptake was comparable between control (Fig. 8D), GMO (Fig. 8E) and Phy treated cells (Fig. 8F). These results suggest that Phy disrupts membrane turnover via macropinocytosis while leaving CME undisturbed. Lipid microdomains within the plasma membrane are thought to be important for macropinocytosis but not CME (see Discussion). The inhibition of macropinocytosis by Phy therefore gives another indication that it disturbs the plasma membrane. The observation that TF uptake was unperturbed in Phy treated cells furthermore suggests that these cells were still viable and that the inhibition of macropinocytosis was not a secondary effect of general toxicity. A similar result was obtained for A549 cells (data not shown).
Cell stress response
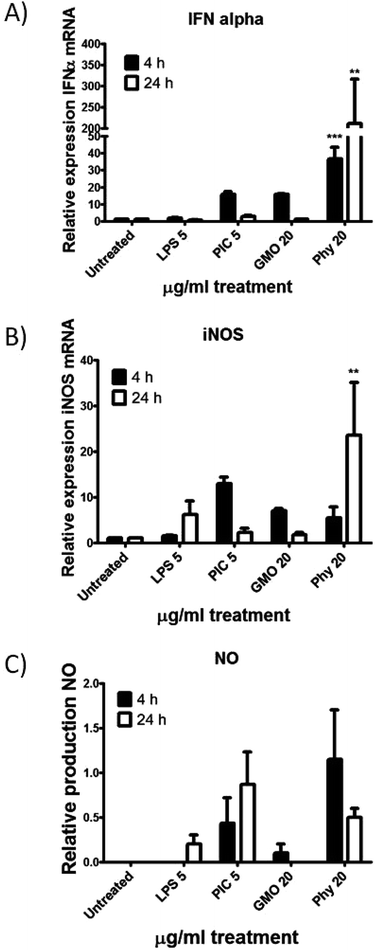
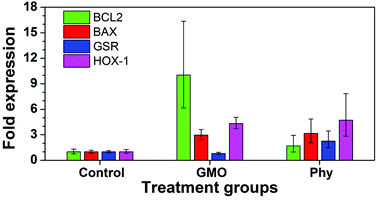
To determine if a pro-inflammatory cell stress response was being triggered in Phy treated cells compared to GMO treated cells, transcriptional activation of two cytokines was assessed. Interferon (INF)-α and inducible nitric oxide synthase (iNOS) are both up regulated in response to pathogens resulting in an inflammatory response. Primary mouse splenocytes were purified and treated with 20 μg ml−1 of either GMO or Phy cubosomes for 4 or 24 h. RNA was purified and quantitative real time PCR (qRT-PCR) was performed. iNOS is responsible for the production of nitric oxide (NO), this was also measured at 24 h. At 4 h, a 36 fold increase in IFN-α mRNA transcription was observed for the Phy treated cells, compared to a 15 fold increase for the GMO and positive control poly I:C samples (PIC) (Fig. 9A). After 24 h both GMO and PIC had returned to untreated levels, whilst Phy treated cells had increased to 200 fold, clearly indicating a significant cell stress response to Phy. At 4 h, a 5 fold increase in iNOS transcription was observed for PIC, GMO and Phy treated cells, however, by 24 h both GMO and PIC had again returned to untreated levels, whilst Phy treated cells transcription increased to 23 fold (Fig. 9B). This corresponded to an increased production of NO in Phy treated cells at 24 h (Fig. 9C).To further investigate the cell stress response to GMO and Phy cubosomes, quantitative real-time PCR was utilized to analyze the mRNA levels of apoptotic (BAX and BCL2), cellular stress (heme oxygenase-1 (HOX-1) and glutathione reductase (GSR)) markers in A549 cells at a cubosome concentration of 20 μg ml−1 for 24 h. The results showed that the mRNA levels of these apoptotic markers were significantly altered in A549 cells due to GMO and Phy nanoparticle exposure. The mRNA level of all four genes was found to be significantly increased in response to Phy nanoparticles (Fig. 10) as compared to the untreated control cells. However, GMO-treated cells showed significant increased level of BCL2, BAX and HOX-1. Interestingly, GSR levels were found to be marginally reduced in the case of GMO and considered insignificant (Fig. 10).
Members of the BCL2 family constitute a group of functionally related proteins that are implicated in the process of cellular apoptosis. While BCL2 is anti-apoptotic in nature, BAX is reported to be a pro-apoptotic protein. Use of certain drugs, chemotherapeutic agents and nanoparticles has been associated with increased cellular apoptosis leading to significant changes in the expression of the BCL2 family of proteins. Following exposure to nanoparticles and chemotherapeutic agents, the ratio of BAX to BCL2 expression has been reported to be a fate determining step for cells.38,39 A lower ratio means a ‘survivor signal’ while a higher ratio has been correlated with cellular apoptosis. A 24 h exposure of GMO and Phy nanoparticles led to a significant increase in the expression of BCL2 and BAX in A549 lung cancer cells (Fig. 10). Cells treated with GMO nanoparticles showed a four-fold increase in BAX expression, which is indicative of a pro-apoptotic response. Interestingly, in order to compensate BAX-induced apoptotic affects, BCL2 levels were also found to be proportionately increased to over ten-fold. This led to the BAX/BCL2 ratio to decrease to less than unity, which is indicative of cellular survival. However, in the case of Phy nanoparticles, a two-fold rise in the BCL2 level was proportionate to the BAX level that showed a five-fold higher expression level leading to a significant increase in the BAX/BCL2 ratio indicative of an apoptotic response. We further measured the levels of oxidative stress-related genes in response to the cubosome nanoparticle incubation. Studies have reported that HOX-1 is a well-known biomarker of oxidative stress, and reported to be induced in animal cells by inorganic mercury and asbestos.40,41 A three-fold increase in HOX-1 and more than a two-fold increase in GSR in the cubosome treated cells might have caused a trigger to cell death in A549 cells resulting in significant cell toxicity due to the Phy nanoparticles. In the case of GMO nanoparticles, although the expression of HOX-1 was significantly increased, the level of GSR remained unchanged. Based on our RT-PCR results it can be concluded that Phy nanoparticles display significantly higher toxic effects as compared to GMO nanoparticles. This toxicity is likely due to increased oxidative stress that leads to cell membrane disruption and an apoptotic response.
Our study indicates that the presence of vesicular structures does not result in increased toxicity, in-fact GMO cubosomes with these structures are less toxic than Phy cubosomes without these structures. One would therefore conclude from our study that the lipid which forms the cubic phase material and the cubic phase itself are the primary cause of cytotoxicity with different bulk lipid chemistries being the important factor. The importance of the cubic phase on cell toxicity is evident. It is clear that the reason for toxicity of these liquid crystal nanoparticles is due to a number of complex factors. Membrane fusion of such systems is dictated by several factors such as nanoparticle size, morphology, charge, defect generation and lipid bilayer permeability. Our study suggests that the cellular response to cubosomes is highly dependent on the bulk lipid chemistry and effectiveness of the polymer stabiliser used to produce the cubosomes thereby moderating the interaction of cubosome nanoparticles with cellular membranes.
To test the hypothesis that cell toxicity was based upon increased uptake of cubic phase forming lipid material, which would result in damage to the cell membrane we devised an assay to study membrane disruption in cultured cells. This assay measured the uptake of the normally membrane-impermeable fluorescent dye lucifer yellow into the cytoplasm of cubosome-treated cells. The results show that a significant amount of damage to the cell membrane is caused by Phy cubosomes compared to the GMO cubosomes. Pogodin et al. have shown via Monte Carlo simulation experiments that the adsorption of amphiphilic nanoparticles onto lipid bilayers can result in membrane destabilization and enhanced permeability of water and small molecules.51 Our lucifer yellow assay has shown that indeed amphiphilic cubosome nanoparticles can disrupt the lipid bilayer of cell membranes with cubosomes made from Phy being particularly effective.
A second assay was utilized by us to determine the ability of the cubosomes to disrupt red blood cell membranes and release haemoglobin. Again Phy cubosomes were able to release significantly more haemoglobin then GMO cubosomes as was alluded to from the lucifer yellow assay we developed. Previous pharmacokinetic studies in rats by us have shown that Myverol cubosomes demonstrated a significantly longer circulation profile than Phy cubosomes. The rapid removal of nanoparticles from circulation is principally a consequence of binding of opsonins and subsequent removal by the reticuloendothelial system (RES).52 Our results indicated that the Myverol cubosomes better avoided recognition by the RES suggesting a more effective steric barrier was provided by the Pluronic F-127 stabiliser in these nanoparticles. This would then result in a greater inhibition of opsonin binding and removal from the bloodstream of GMO cubosomes. This is consistent with past observations that Myverol nanoparticle dispersions demonstrated improved colloidal stability compared to phytantriol dispersions. This has been proposed to be linked to stronger interactions of the Pluronic F-127 stabilizer with the Myverol nanoparticles.47 Therefore, one hypothesis to explain the observation that GMO cubosomes are less toxic than Phy cubosomes is that upon contact with the cell membrane, the bulk lipid in Phy cubosomes is able to interact with the cell membrane to a greater extent (in part due to less effective Pluronic F-127 stabilization). Phy would therefore be more likely to disrupt the cell membrane to a greater extent when compared to GMO.
An important recent study of note is that of Barauskas et al.23 who reported an investigation of the interaction of various cubic an hexagonal phase forming lipids with model cell membranes. They have shown that cubic phase nanoparticles (including those made from GMO) are significantly more effective at disrupting and mixing with model lipid bilayers than lamellar and hexagonal phase nanoparticles. When GMO cubosomes were incubated with rat whole blood they saw significant haemolysis of red blood cells while no effect was seen with hexagonal phase nanoparticles (hexosomes). They attributed this haemolytic activity of GMO cubosomes to be due to its ability to destabilize cell membranes thereby causing phase segregation of lipid bilayers within cell membranes introducing pore structures resulting in leakage of cellular content. More recently Tilley et al.53 have shown that phytantriol cubosomes can transfer their lipid material between lamellar and hexagonal phase nanoparticles in solution. This process occurs via compositional ripening. The driving force for mixing of the cubic phase nanoparticles with lipid membranes is due to the entropy of mixing with the system attempting to achieve a more thermodynamically favorable state by attaining the same composition. It was also found that when particles of similar lipophilicities were mixed, lipid transfer was limited and did not achieve completion. A phase change between cubic nanostructures required to achieve complete mixing was proposed to provide a barrier to further compositional ripening. It has been shown that the direction of lipid exchange is dependent on the chemical potential between the two lipid systems. Other researchers have found similar results in equivalent systems.54–57
In our study and that of Shen et al.25 there is clear evidence of transfer of the R18 dye from the cubosomes produced (particularly those of Phy) into cellular structures which supports the observations of other researchers when studying mixing of bulk lipids between different lyotropic liquid crystal phase nanoparticles and model cell membranes. The exact location of fluorescence from the rhodamine lipid (R18) cannot be pinpointed but it is known that other rhodamine dyes tend to associate with mitochondria in living cells.58 In fact, rhodamine dyes can be used to target mitochondria therefore one might imagine that a percentage of the fluorescence we have observed in our study will be a result of the R18 dye associating with mitochondria.
To conclude, we have shown that important differences exist between the toxicity of GMO and Phy, two very similar and commonly used cubic phase lipid nanoparticles. Phy is significantly more toxic, and we provide evidence that this is due to a disruption of membrane integrity, oxidative stress and possibly the order and composition of plasma membrane lipids. These results have important implications for the development of cubic phase lipid nanoparticles for in vivo use.
Acknowledgements
RS and VB would like to acknowledge Ian Potter Foundation for generous instrument support.References
- X. Mulet, B. J. Boyd and C. J. Drummond, J. Colloid Interface Sci., 2013, 408, 117 CrossRef PubMed.
- X. Mulet, C. E. Conn, C. Fong, M. Moghaddam, D. F. Kennedy and C. J. Drummond, Acc. Chem. Res., 2013, 46, 1497 CrossRef CAS PubMed.
- V. Jain, N. K. Swarnakar, P. R. Mishra, A. Verma, A. Kaul, A. K. Mishra and N. K. Jain, Biomaterials, 2012, 33, 7206 CrossRef CAS PubMed.
- T.-H. Nguyen, T. Hanley, C. J. H. Porter, I. Larson and B. J. Boyd, J. Pharm. Pharmacol., 2010, 62, 844 CAS.
- T.-H. Nguyen, T. Hanley, C. J. H. Porter, I. Larson and B. J. Boyd, J. Pharm. Pharmacol., 2010, 62, 856 CAS.
- P. T. Spicer, Curr. Opin. Colloid Interface Sci., 2005, 10, 274 CrossRef CAS PubMed.
- M. Johnsson, J. Barauskas, A. Norlin and F. Tiberg, J. Nanosci. Nanotechnol., 2006, 6, 3017 CrossRef CAS PubMed.
- K. T. Love, K. P. Mahon, C. G. Levins, K. A. Whitehead, W. Querbes, J. R. Dorkin, J. Qin, W. Cantley, L. L. Qin, T. Racie, M. Frank-Kamenetsky, K. N. Yip, R. Alvarez, D. W. Y. Sah, A. de Fougerolles, K. Fitzgerald, V. Koteliansky, A. Akinc, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1864 CrossRef CAS PubMed.
- N. Tri-Hung, T. Hanley, C. J. H. Porter and B. J. Boyd, J. Controlled Release, 2011, 153, 180 CrossRef PubMed.
- J. Barauskas, M. Johnsson, F. Joabsson and F. Tiberg, Langmuir, 2005, 21, 2569 CrossRef CAS PubMed.
- L. Sagalowicz, M. E. Leser, H. J. Watzke and M. Michel, Trends Food Sci. Technol., 2006, 17, 204 CrossRef CAS PubMed.
- B. W. Muir, D. P. Acharya, D. F. Kennedy, X. Mulet, R. A. Evans, S. M. Pereira, K. L. Wark, B. J. Boyd, T. H. Nguyen, T. M. Hinton, L. J. Waddington, N. Kirby, D. K. Wright, H. X. Wang, G. E. Egan and B. A. Moffat, Biomaterials, 2012, 33, 2723 CrossRef CAS PubMed.
- L. Gan, S. Han, J. Shen, J. Zhu, C. Zhu, X. Zhang and Y. Gan, Int. J. Pharm., 2010, 396, 179 CrossRef CAS PubMed.
- S. Han, J.-q. Shen, Y. Gan, H.-m. Geng, X.-x. Zhang, C.-l. Zhu and L. Gan, Acta Pharmacol. Sin., 2010, 31, 990 CrossRef CAS PubMed.
- J. Barauskas, M. Johnsson, F. Johnson and F. Tiberg, Langmuir, 2005, 21, 2569 CrossRef CAS PubMed.
- M. Johnsson, Y. Lam, J. Barauskas and F. Tiberg, Langmuir, 2005, 21, 5159 CrossRef CAS PubMed.
- X. Gong, M. J. Moghaddam, S. M. Sagnella, C. E. Conn, X. Mulet, S. J. Danon, L. J. Waddington and C. J. Drummond, Soft Matter, 2011, 7, 5764 RSC.
- C. Cervin, M. Tinzl, M. Johnsson, P. A. Abrahamsson, F. Tiberg and N. Dizeyi, Eur. J. Pharm. Sci., 2010, 41, 369 CrossRef CAS PubMed.
- W. Leesajakul, M. Nakano, A. Taniguchi and T. Handa, Colloids Surf., B, 2004, 34, 253 CrossRef CAS PubMed.
- S. M. Sagnella, X. Gong, M. J. Moghaddam, C. E. Conn, K. Kimpton, L. J. Waddington, I. Krodkiewska and C. J. Drummond, Nanoscale, 2011, 3, 919 RSC.
- D. P. Acharya, B. A. Moffat, A. Polyzos, L. Waddington, G. Coia, D. K. Wright, H. X. Wang, G. F. Egan, B. W. Muir and P. G. Hartley, RSC Adv., 2012, 2, 6655 RSC.
- A. Angelova, B. Angelov, R. Mutafchieva, S. Lesieur and P. Couvreur, Acc. Chem. Res., 2011, 44, 147 CrossRef CAS PubMed.
- J. Barauskas, C. Cervin, M. Jankunec, M. Spandyreva, K. Ribokaite, F. Tiberg and M. Johnsson, Int. J. Pharm., 2010, 391, 284 CrossRef CAS PubMed.
- S. Murgia, A. M. Falchi, M. Mano, S. Lampis, R. Angius, A. M. Carnerup, J. Schmidt, G. Diaz, M. Giacca, Y. Talmon and M. Monduzzi, J. Phys. Chem. B, 2010, 114, 3518 CrossRef CAS PubMed.
- H. H. Shen, J. G. Crowston, F. Huber, S. Saubern, K. M. McLean and P. G. Hartley, Biomaterials, 2010, 31, 9473 CrossRef CAS PubMed.
- T. Yu, A. Malugin and H. Ghandehari, ACS Nano, 2011, 5, 5717 CrossRef CAS PubMed.
- G. Oberdorster, V. Stone and K. Donaldson, Nanotoxicology, 2007, 1, 2 CrossRef CAS.
- K. Unfried, C. Albrecht, L. O. Klotz, A. VonMikecz, S. Grether-Beck and R. P. F. Schins, Nanotoxicology, 2007, 1, 52 CrossRef CAS.
- J. M. Zook, R. I. MacCuspie, L. E. Locascio, M. D. Halter and J. T. Elliott, Nanotoxicology, 2011, 5, 517 CrossRef CAS PubMed.
- E. Frohlich, C. Meindl, E. Roblegg, A. Griesbacher and T. R. Pieber, Nanotoxicology, 2012, 6, 424 CrossRef PubMed.
- J. Jiang, G. Oberdorster, A. Elder, R. Gelein, P. Mercer and P. Biswas, Nanotoxicology, 2008, 2, 33 CrossRef CAS PubMed.
- Y. Gao, R. F. Yang, Z. W. Zhang, L. L. Chen, Z. Y. Sun and Y. P. Li, Nanotoxicology, 2011, 5, 636 CrossRef CAS PubMed.
- G. Zhen, T. M. Hinton, B. W. Muir, S. Shi, M. Tizard, K. M. McLean, P. G. Hartley and P. Gunatillake, Mol. Pharm., 2012, 9, 2450 CrossRef CAS PubMed.
- P. Chomczynski and N. Sacchi, Anal. Biochem., 1987, 162, 156 CrossRef CAS.
- S. Lienenklaus, R. Walisko, A. T. Boekhorst, T. May, C. Samuelsson, T. Michiels and S. Weiss, J. Interferon Cytokine Res., 2008, 28, 653 CrossRef CAS PubMed.
- S. Burggraaf, J. Bingham, J. Payne, W. G. Kimpton, J. W. Lowenthal and A. G. D. Bean, PLoS One, 2011, 6 Search PubMed.
- M. H. Elfouly, J. E. Trosko and C. C. Chang, Exp. Cell Res., 1987, 168, 422 CrossRef CAS.
- G. Xu, Z. Gong, W. Yu, L. Gao, S. He and Z. Qian, Basic Clin. Pharmacol. Toxicol., 2007, 100, 31 CrossRef CAS PubMed.
- M. E. Selim and A. A. Hendi, Asian Pac. J. Cancer Prev., 2012, 13, 1617 CrossRef.
- E.-J. Park, J. Choi, Y.-K. Park and K. Park, Toxicology, 2008, 245, 90 CrossRef CAS PubMed.
- H. Nagatomo, Y. Morimoto, A. Ogami, M. Hirohashi, T. Oyabu, K. Kuroda, T. Higashi and I. Tanaka, Inhal. Toxicol., 2007, 19, 317 CrossRef CAS PubMed.
- A. V. Kabanov and V. Y. Alakhov, Amphiphilic Block Copolymers: Self-Assembly and Applications, 2000, p. 347 Search PubMed.
- F. Liu, J. P. Yang, L. Huang and D. X. Liu, Pharm. Res., 1996, 13, 1642 CrossRef CAS.
- N. S. Melik-Nubarov, O. O. Pomaz, T. Y. Dorodnych, G. A. Badun, A. L. Ksenofontov, O. B. Schemchukova and S. A. Arzhakov, FEBS Lett., 1999, 446, 194 CrossRef CAS.
- C. Leal, N. F. Bouxsein, K. K. Ewert and C. R. Safinya, J. Am. Chem. Soc., 2010, 132, 16841 CrossRef CAS PubMed.
- C. Leal, K. K. Ewert, R. S. Shirazi, N. F. Bouxsein and C. R. Safinya, Langmuir, 2011, 27, 7691 CrossRef CAS PubMed.
- Y.-D. Dong, I. Larson, T. J. Bames, C. A. Prestidge and B. J. Boyd, ACS Appl. Mater. Interfaces, 2011, 3, 1771 CAS.
- J. Y. T. Chong, X. Mulet, L. J. Waddington, B. J. Boyd and C. J. Drummond, Langmuir, 2012, 28, 9223 CrossRef CAS PubMed.
- A. J. Tilley, C. J. Drummond and B. J. Boyd, J. Colloid Interface Sci., 2013, 392, 288 CrossRef CAS PubMed.
- H.-H. Shen, P. G. Hartley, M. James, A. Nelson, H. Defendi and K. M. McLean, Soft Matter, 2011, 7, 8041 RSC.
- S. Pogodin, M. Werner, J.-U. Sommer and V. A. Baulin, ACS Nano, 2012, 6, 10555 CrossRef CAS PubMed.
- L. M. Kaminskas, V. M. McLeod, C. J. H. Porter and B. J. Boyd, J. Pharm. Sci., 2011, 100, 5069 CrossRef CAS PubMed.
- A. Tilley, Y.-D. Dong, H. Amenitsch, M. Rappolt and B. J. Boyd, Phys. Chem. Chem. Phys., 2011, 13, 3026 RSC.
- C. Moitzi, S. Guillot, G. Fritz, S. Salentinig and O. Glatter, Adv. Mater., 2007, 19, 1352 CrossRef CAS.
- A. Salonen, C. Moitzi, S. Salentinig and O. Glatter, Langmuir, 2010, 26, 10670 CrossRef CAS PubMed.
- P. Vandoolaeghe, J. Barauskas, M. Johnsson, F. Tiberg and T. Nylander, Langmuir, 2009, 25, 3999 CrossRef CAS.
- P. Vandoolaeghe, A. R. Rennie, R. A. Campbell and T. Nylander, Langmuir, 2009, 25, 4009 CrossRef CAS.
- L. V. Johnson, M. L. Walsh and L. B. Chen, Proc. Natl. Acad. Sci. U. S. A., 1980, 77, 990 CrossRef CAS.
- H. Hillaireau and P. Couvreur, Cell. Mol. Life Sci., 2009, 66, 2873 CrossRef CAS PubMed.
- S. Grimmer, B. van Deurs and K. Sandvig, J. Cell Sci., 2002, 115, 2953 CAS.
- M. F. Hanzal-Bayer and J. F. Hancock, FEBS Lett., 2007, 581, 2098 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3tx50075f |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2014 |