Open Access Article
Open Access ArticlePreventing bacterial colonization using colloidal crystals†
Mehdi
Kargar
a,
Amy
Pruden
b and
William A.
Ducker
*c
aDepartment of Mechanical Engineering, Virginia Tech, Blacksburg, VA, USA. E-mail: mehdik@vt.edu; Fax: +1-540-231-5022; Tel: +1-540-231-7869
bVia Department of Civil and Environmental Engineering, Virginia Tech, Blacksburg, VA, USA. E-mail: apruden@vt.educ; Fax: +1-540-231-7816; Tel: +1-540-2313980
cDepartment of Chemical Engineering, Virginia Tech, Blacksburg, VA, USA. E-mail: wducker@vt.edu; Fax: +1-540-231-5022; Tel: +1-540-231-7869
First published on 2nd July 2014
Abstract
We investigated the adhesion and colony formation of Pseudomonas aeruginosa PAO1 on a solid coated in close-packed spheres of polystyrene. The objective was to determine the effect of surface topography on the early stages of biofilm formation. Solids were pretreated with serum and then exposed to bacteria under low shear for one day in a center for disease control biofilm reactor. Whereas flat sheets are covered in large colonies after one day, a close-packed layer of 630–1550 nm monodisperse spheres prevents colony formation. Moreover, the film of spheres reduces the density of P. aeruginosa adhered to the solid by an average of 80%. Our data show that when P. aeruginosa adheres to the spheres, the distribution is not random. For 630 nm and larger particles, P. aeruginosa tends to position its body in a 2-fold site. We rationalize the selectivity on the basis of energy minimization for adhesion: sites differ in the deformation needed to achieve a given contact area. We rationalize the inhibition of colonization by the 630–1550 nm spheres in terms of the lack of adjacent favorable positions for bacteria. A close-packed layer of polystyrene spheres also delays colony formation on a medical-grade stainless-steel needle over a period of one day. This suggests that a colloidal crystal approach to biofilm inhibition might be applicable to a variety of materials and geometries.
1. Introduction
Bacterial infections are one of the leading causes of human misery, and hospital acquired infections alone are the fourth most common cause of death in the US.1 About 65% of these deaths are caused by infections invading the surface of implants and medical devices.2 These infections begin with the adhesion of microorganisms to surfaces, continue with the formation of microbial colonies and lead to the formation of biofilms.22 In addition to misery and death, these infections are estimated to cost the US approximately $5 billion annually.2The most common medical treatment of device and implant infections is long-term systemic treatment with antibiotics.3 Antibiotic treatments sometimes fail because of the development of biofilms, which are less responsive to antibiotic treatments,4 or the presence antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA).5 Antibiotic treatments frequently have broad systemic effects on the body, including killing beneficial microbes.6
An alternative strategy for reducing the incidence of these infections is to alter the properties of the medical surfaces to delay colonization by bacteria. Chemical modifications have been widely studied, to either delay adhesion of microbes to the surfaces or kill them upon contact.7 Surface topography is another property of the surface that can complement the effects of chemistry to reduce fouling8 further.
Antifouling surfaces with topographical features are observed widely in nature,9–12 and this has inspired the preparation of artificial surface patterning to prevent the adhesion of bacteria. For example, Chung et al.13 prepared a surface that mimicked the topography of shark skin. This surface contained protrusions 4–16 μm in length, 2 μm in width and 3μm in height, spaced by 2 μm. Chung et al. showed that although these features did not prevent adhesion of bacteria, the formation of biofilms was greatly delayed. The authors attributed this effect to the “physical obstacle” presented by the protrusions.
A number of other studies have examined the effect of topography on bacterial interactions with surfaces.13–21 For example, Hochbaum and Aizenberg22 studied interactions of bacteria with submicron-size pillars, and concluded that when interacting with the spacing between the pillar, each cell selected an alignment that provided it with greater contact area with the surface. Kargar et al.17 investigated the adhesion of P. aeruginosa to fibrous surfaces at a single cell level. They showed that P. aeruginosa selectively binds to specific locations of the fibrous surfaces, and gave insights into the action of local surface curvature on hindering microbial adhesion.
One of the major barriers to advancing topographical-based antifouling technologies is fabricating the structure. Well-organized structures within the size range of bacteria (submicron-micron) are needed over the exposed area of a medical device. Most techniques used in the past are constrained by a limited range of suitable materials and/or the requirement of sophisticated facilities, which adds greatly to the expense of manufacturing. The purpose of this study was to examine the use of an adsorbed monolayer of close-packed spheres as a topographical inhibitor of bacterial adhesion and colony formation.
A regular arrangement of colloidal particles is also called a “colloidal crystal”, and in this paper we use the term colloidal crystals to refer to a monolayer of close packed spheres. The principal advantages of colloidal crystal to prevent microbial colonization are: (1) they can be used to coat a broad range of surfaces, regardless of their chemistry;23,24 (2) they can coat surfaces with complex geometries, for example curved and non-planar surfaces;23–25 (3) they can be used to produce a broad range of feature sizes (larger than 50 nm);14 (4) they can be produced on large areas at low cost in a short time without the requirement of sophisticated facilities such as a clean room;25 and (5) they can provide surfaces with well-defined curvatures in nano-micron scale.26 Here we examine the effects of polystyrene colloidal crystal coatings (diameter 200 nm–1600 nm) on bacterial colonization of polystyrene (PS). All PS samples were exposed to fetal bovine serum before bacterial experiments. This leaves an adsorbed layer of protein, which is common for surfaces in biological systems. We use Pseudomonas aeruginosa, a rod shape, Gram-negative and motile bacteria as our model bacterium, it is an opportunistic human pathogen that exists in biofilms in hospital environments.27
2. Experimental
2.1. Chemicals
The following chemicals and regents were all obtained from Fisher scientific (Pittsburg, PA): acetone (HPLC grade), sulfuric acid (18 M), tryptic soy broth (TSB) powder, tryptic soy agar (TSA) powder and Hyclone fetal bovine serum (FBS). TSA plates were prepared by adding 1 liter of DI water to 40 g of TSA. TSB media was prepared by adding 1 liter of water to 30 g of TSB powder. The other chemicals used in the experiments are: hydrogen peroxide, 30% in water (LabChem Inc., Zelienople, PA), USP grade 200 proof ethanol (Decon Laboratories, King of Prussia, PA) and 10% EM grade glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) sodium dodecyl sulfate (SDS) and potassium nitrate were purchased from Sigma-Aldrich (St. Louis, MO). All PS spheres have been purchased from Bangs laboratory incorporation (Fishers, IN). The flat sheets of PS film that were used as control flat surfaces were purchased from Goodfellow (Coraopolis, PA). Deionized (DI) water used in the experiments had resistivity 18.2 MΩ cm at 25 °C. Diluted TSB, ethanol and glutaraldehyde have made by mixing DI water and the original chemical with the appropriate ratio. L7007 live dead assay kit was purchased from Invitrogen (Grand Island, NY).2.2. Manufacturing the colloidal crystal monolayers
Several methods for manufacturing colloidal crystal films are described in the literature.28 Our films were prepared by first depositing particles at the liquid–gas interface, and then pulling a solid from within the solution out into the air through the interface (emerging).23–25,29 Here we will refer to the solid underneath the particles as the “substrate” and the colloidal crystal film plus substrate that is subsequently used for bacterial testing as the “sample”. We have adopted the specific procedure from Lu and Zhou.29 Briefly, the particles were re-suspended in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethanol–DI water solution and deposited at the water/air interface by flowing the particle suspension down a glass slide. The slides were cleaned using consecutive washes with acetone, ethanol, DI water followed by nitrogen drying and cleaning by Piranha solution (1/3 H2O2–H2SO4). The particles were then deposited on the substrate by emerging the substrate though the interfacial particles. In most cases the substrate was a flat sheet of polystyrene. As recommended by Lu and Zhou,29 we have added a small amount of SDS to the particles to facilitate the formation of the closed pack structures. After formation of the colloidal crystal, we heated the sample at 95 °C for 110 min. This heating close to the glass transition temperature increases the area of contacts and thus strengthens bonds between particles and between the particles and the substrate.
1 ethanol–DI water solution and deposited at the water/air interface by flowing the particle suspension down a glass slide. The slides were cleaned using consecutive washes with acetone, ethanol, DI water followed by nitrogen drying and cleaning by Piranha solution (1/3 H2O2–H2SO4). The particles were then deposited on the substrate by emerging the substrate though the interfacial particles. In most cases the substrate was a flat sheet of polystyrene. As recommended by Lu and Zhou,29 we have added a small amount of SDS to the particles to facilitate the formation of the closed pack structures. After formation of the colloidal crystal, we heated the sample at 95 °C for 110 min. This heating close to the glass transition temperature increases the area of contacts and thus strengthens bonds between particles and between the particles and the substrate.
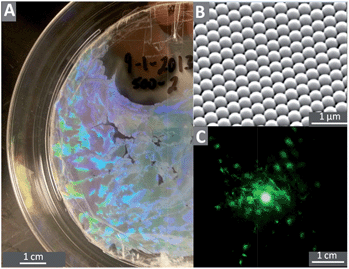
This method enabled us to manufacture colloidal crystals from various particle sizes but the reproducibility was low. Our original substrates were 100 mm circles of a 0.125 mm thick PS film. After deposition, we cut 1.5 × 0.5 mm2 rectangular samples from the colloidal crystal for our biological test assay. The quality of the film was always characterized using a combination of the diffraction patterns from a laser, and scanning electron microscopy (SEM) imaging.
2.3. Characterization of substrates, spheres and samples
XPS measurements were performed on a Phi Quantera SXM with a monochromatic Al Kα X-ray source (1486 eV). All of the XPS measurements were obtained at a 45° takeoff angle from a 200 μm diameter spot. The wide energy scans were acquired with a 280 eV pass energy in 15 sweeps at 1 eV step size and narrow energy scans were acquired at pass energy 26 eV with 10–40 sweeps depending on the element at 0.1 eV step size.Zeta potential measurements of colloidal spheres were performed with a Zetasizer Nano ZS (Malvern, Worcestershire, United Kingdom). The zeta potential was obtained from the mobility using the appropriate correction term for the ratio between the radius and Debye-length.30 The zeta potential of the flat PS film and particle-coated surfaces was determined using a SurPASS Electrokinetic Analyzer (Anton Paar GmbH, Graz, Austria). The surfaces were washed with water, then ethanol, and then dried in stream of pure nitrogen. The solvent for all zeta potential measurements was 0.05 mM KNO3 solution.
The roughness of the flat PS substrates was measured using atomic force microscopy (AFM) (MFP3D, Asylum Research, Santa Barbara, CA) using ORC8 cantilevers (Bruker, Camarillo, CA) with a nominal spring constant of 0.7 N m−1 in contact mode.
2.4. Preparation of samples for biological assays
All samples were washed with DI water at 37 °C in a rotary shaker (100 rpm) for 1 hour and stored in Petri dishes. Before biological tests, the samples were washed with 100% ethanol and sterilized by soaking in 70% ethanol following by three washes with autoclaved DI water. In order to mimic the conditioning film that samples will experience in a biological environment all samples were submerged under 2 ml FBS in wells of 12 well plates and incubated at 37 °C overnight, following a protocol for testing catheters.31 Negative control experiments (i.e. with no bacteria) were conducted by transferring some of the FBS-treated samples to a new 12 well plate and incubating them under sterile TSB for a day. Samples that were coated in FBS had a water contact angle <10°.2.5. Bacterial culturing and retention assay
Pseudomonas aeruginosa (ATCCR 47085ࡊ), PAO1, was used as the model microorganism in a Center for Disease Control (CDC) reactor (BioSurface Technologies Co., Bozeman, MO). We followed the standard protocol developed by Goeres et al.32 and recommended by the manufacturer of the biofilm reactor with some minor modifications.A 1.5% TSA plate was streaked using bacteria from −80 °C stock and incubated overnight at 37 °C. 100 ml of autoclaved culture media (1% TSB) was transferred to a sterile 300 ml flask, inoculated using a single colony from the TSA plate and incubated for 24 hours at 37 °C on a rotary shaker (170–180 rpm). These conditions consistently resulted in OD600 ≈ 0.06. The samples were mounted on the rods of CDC reactor, 323.3 ml of 1% TSB were added, and then 1.7 ml of bacterial culture was used to inoculate the reactor. A dynamic retention assay was conducted for 24 hours at 37 °C, in which the baffle of the reactor was stirring at 50 RPM. To estimate the shear stress due to rotation of the baffle at the surface of the samples, we have followed the method of Buckingham-Meyer et al.33 and used the equations for the shear forces between concentric cylinders where the inner cylinder rotates. This suggests that 50 RPM baffle speed results in a shear stress of about 0.004 N m−2, which is low compared to the shear rate in other studies.20
2.6. Imaging of the samples using scanning electron microscope (SEM)
The results presented here were collected from four different experiments. We define a different experiment to be one done using different substrates, different microbial stock, and on a different day. After each experiment, all samples were washed once with autoclaved DI water to remove the loosely attached cells. Then the cells were fixed in 2.5% glutaraldehyde at 4 °C and washed with DI water to remove the glutaraldehyde. Samples were then dehydrated using serial ethanol washes and dried for a day at room temperature. All samples were gold coated and then imaged at high vacuum using a scanning electron microscope (FEI Quanta 600 FEG, Hillsboro, OR). The images presented for the steel catheter were captured by a different microscope (LEO 1550 field-emission SEM, Carl Zeiss, Thornwood, NY).2.7. Imaging of the samples using light microscopy
Light microscopy was used to image the cells in the hydrated state (under water), and after dehydration. Samples were washed with autoclaved DI water (as for SEM) and then fixed in 2.5% glutaraldehyde at 4 °C. For fluorescent microscopy, the cells stained with a mixture of propidium iodide and SYTO®9 nucleic acid stain. For all imaging, samples were attached to the bottom of glass-bottom Petri dishes using PDMS and imaged using an Olympus 1X81microscope and Slidebook 5 software.2.8. Data analysis and statistics
SEM images were analysed quantitatively. Only the 0.5 × 0.5 mm2 square shaped area in the middle of the 1.5 × 0.5 mm2 area of the sample was used, so as to avoid edge effects. A standard grid containing the positions for counting was placed over a low magnification image (35×) in which the bacteria were too small to be imaged. Images were then captured at each of 20 positions on this standard grid, and the number of bacteria per unit area was obtained by manual counting and then averaged. Cell counting was assisted by ImageJ software,34 and the convergence of the counting results was monitored during the counting process. For most particle samples, convergence occurred after counting 8–10 samples, but for the flat surfaces further counting was required. The magnification of the images used in counting process was 3500× for flat samples and 6000× for colloidal crystals.3. Results
3.1. Characterization of flat and colloidal-crystal-coated surfaces
| Diameter (nm) | Zeta potential (mV) | St. De. (mV) |
|---|---|---|
| 220 | −77 | 7 |
| 450 | −73 | 11 |
| 630 | −79 | 8 |
| 830 | −88 | 8 |
| 925 | −89 | 8 |
| 1550 | −94 | 13 |
| Flat | −54 | 4 |
There was only a small variation in the zeta potential of the particles (∼20%), but the flat sheet had a less negative potential than the particles, despite the similarity in the chemistry as measured by XPS. This difference may be explained in part due to different techniques used (electrophoresis vs. streaming potential). After noting this possible difference, we created additional flat samples in which the colloidal crystal on a flat glass substrates was sintered by heat treatment at 120 °C (higher than the glass transition temperature of PS) for 12 days followed by heat treatment at 140 °C for one day, by which time the spheres Ostwald ripened into flat sheets. We refer to these as flattened samples. When comparing the flattened samples to a colloidal crystal, we were even more confident that the samples had the same chemistry. Further, we noted that the flattened samples and the flat samples had the same water contact angle of 91°.
3.2. Effect of sphere coating on bacterial adhesion and colony formation
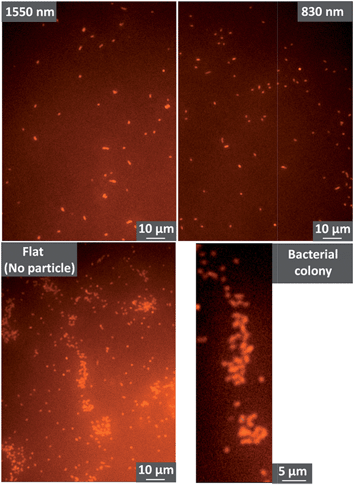 | ||
| Fig. 2 Fluorescence images showing P. aeruginosa on polystyrene colloidal crystals (1550 nm and 830 nm diameter spheres) and on a flat polystyrene sheet. Further images are shown in Fig. S1 in the ESI.† | ||
To enable higher resolution imaging of colloidal crystals and the bacteria, we also performed SEM imaging. Sample preparation for SEM requires dehydration and drying of the bacteria, so we first tested whether our SEM sample preparation altered the distribution or number of cells on the solid. Previous work has shown that the passage of air bubbles in water can alter surface distributions of bacteria.35 As shown in Fig. S2 and S3 in the ESI,† exchange of the solution with ethanol, then air-drying affected neither the number density, nor the arrangement of the P. aeruginosa on the colloidal crystal and flat surfaces. Likewise Fig. S4 shows that the passage of the ethanol–air interface across a particle does not change the position of the bacterium.
Knowing that the sample preparation does not affect the distribution of bacteria, we can use SEM to examine the effect of the colloidal crystal in more detail. The SEM image in Fig. 3 shows large colonies on flat surfaces, with the same distribution as in the optical images.
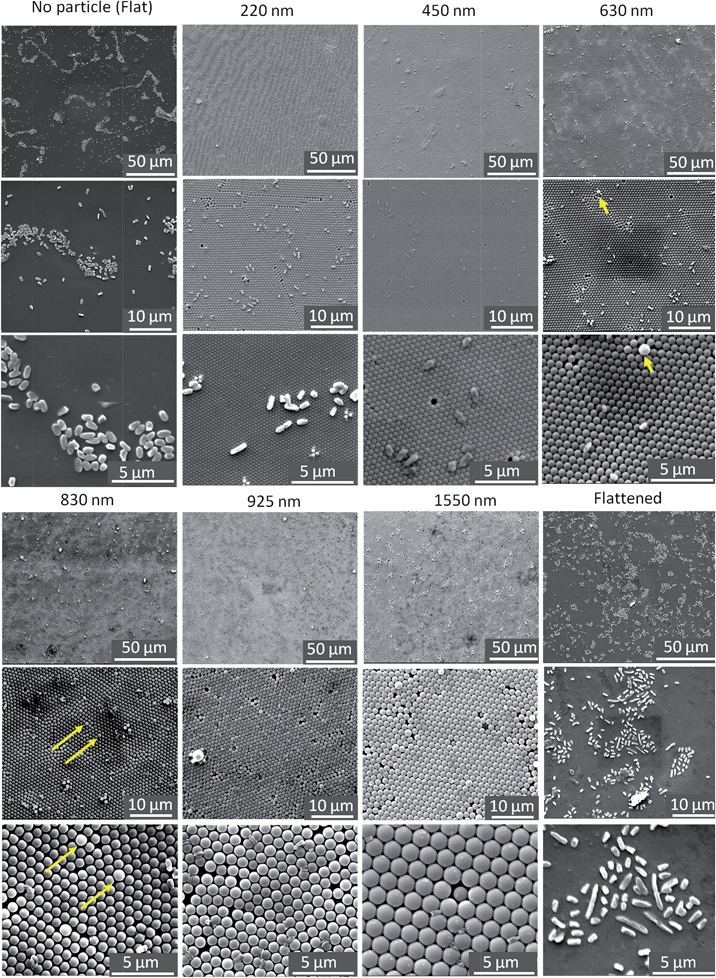
Our first objective is to examine how the spheres affect colony formation. A colony is a collection of cells in which there is close contact,36 but there is no universal definition of the number of cells required for a colony. For the purpose of quantification here, we will define a colony as a collection of bacteria which has more than 10 cells. Table 2 shows three quantities for both flat and colloidal crystals: (1) colony density: the number of colonies per 1 mm2 of the projected area of a sample, (2) colony size: the average number of cells in a colony, and (3) % in colony: the number of bacteria in colonies divided by the total number of cells available on the surfaces.
| Diameter | Flat | 220 nm | 450 nm | 630–1550 nm |
|---|---|---|---|---|
| Colony density (colony per mm2) | 732 | 446 | 38 | 0 |
| Colony size (cells per colony) | 56 | 20 | 12 | 0 |
| % In colony | 83 | 22 | 5 | 0 |
Our results indicate that adding smaller particles (220 nm) to the surface will reduce both size of the colony and the density of the colonies. An even smaller density of even smaller colonies was observed on the 450 nm colloidal crystals; only 5% were involved in colonies. The average colony size was only 12 and if we had made our arbitrary definition of colony to be 16 cells, then there would be zero colonies on the 450 nm sample. No colonies were observed on crystals formed from the larger spheres. Thus all the sphere sizes reduced the number of colonies.
Experiments using flattened samples typically produced similar results as those on the flat polystyrene shown in Fig. 3, i.e. large colonies formed. Thus, comparing the colloidal crystal samples to the flattened samples (which were formed from exactly the same material) we conclude that the topography prevented colony formation on the colloidal crystals. Comparing the flattened samples to the flat sheet samples, both allowed the formation of colonies, thus variation in zeta potential of the polystyrene under the FBS-deposited layer was not the determining factor for colony formation in this work.
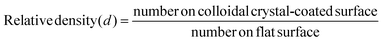 | (1) |
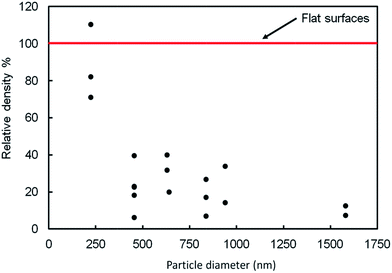
We conducted a one way analysis of variance (ANOVA) followed by a Tukey test to determine whether there was a significant difference (p ≤ 0.05) between the relative densities of bacterial colonies at different particle diameters. The ANOVA test showed that the relative density of bacteria adhered to the 220 nm particle film was not significantly different from the density on the flat surface. The diameter did not have a significant effect on the relative densities of bacterial colonies for the particle size range 450–1550 nm. However, we did observe a significant difference between the number of bacteria per unit area on the flat surface and any sample using spheres in the size range 450–1550 mn: the number of bacteria was reduced by more than 80% on these larger-diameter colloidal crystals.
In summary, the 450–1550 nm particles prevented the formation of large colonies and reduced the number of bacteria by about 80% compared to the flat surface. The 250 nm particles did not reduce the number of bacteria, but still inhibited the formation of large colonies.
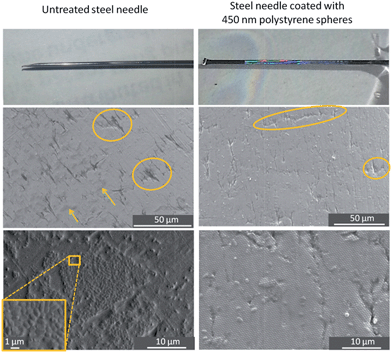
A colloidal crystal coating was prepared on the needle using the same method as for the flat polystyrene substrate. The optical photograph showing an opalescent film on the needle in Fig. 6 signifies that a colloidal crystal (450 nm particles) can be deposited on a curved material that is not polystyrene, and SEM imaging shows that the particles are arranged in much the same way as on a flat sheet of polystyrene. The SEM pictures in Fig. 6 demonstrate that colony formation is dramatically hindered on the colloidal-crystal-coated needle compared to the untreated needle.
4. Discussion
4.1. Concepts of selective adhesion
The SEM images show that the bacteria are not randomly distributed across the samples: specific locations have a greater density of occupation. Previous work suggests that both strain specificity and motility are important factors in selectivity.12,17,19,20,22,37–41 With respect to motility, Scardino et al.40 compared the interactions of motile and non-motile marine organisms with topographically engineered surfaces and reported that whereas the number of adhered motile cells depended on the topography, the number of adhered non-motile cells did not. Kargar et al.37 observed similar behavior for P. aeruginosa PAO1: wild type P. aeruginosa selectively attached to specific binding sites on fibrous surfaces whereas the non-motile pila mutants, (lacking type IV pili, used for surface motility) lacked selectivity. Furthermore, P. aeruginosa, has been observed to show selectivity in its alignment relative to topographical features.13,16,42The preference for particular sites on the colloidal crystal observed in this work suggests that P. aeruginosa does discriminate among different sites. This discrimination should have two aspects: (1) the ability to move between two sites and (2) differences between sites that affect the occupancy of the site by the bacterium. The ability to move between sites can be provided by the motile elements such as pili or flagella, or by passive action such as diffusion or fluid flow (provided by stirring in our experiments).
Given the opportunity to explore, the second component of selective adhesion is discrimination among different sites. Bacteria are complex: they have proteinaceous features in their membrane which contribute in their adhesion to the surfaces, secrete polymers to enhance adhesion,43 and they can communicate.44 All these factors may be important, but here we focus on the physical properties of deformation and adhesion and the relationship to topography. Kargar et al.17 explained selective adhesion of P. aeruginosa to fiber-coated surfaces based on a balance between adhesion energy and deformation energy, following the work on vesicle adhesion described by Seifert and Lipowsky.45 In the same vein, we make an assumption: a favorable adsorption site is where a large area of contact can be achieved between the bacterium and the solid. The logic behind the assumption is that a large area enables a strong adhesion if adhesion is proportional to the area of contact. At each point where a bacterium contacts a solid, the bacterium may deform or secrete compounds to increase the contact area, but each of these costs energy. The important aspect of our colloidal crystals is the bacterium can only contact a sphere, and, each sphere curves away from the bacterium, meaning that, as the area of the contact grows, the distance of deformation grows, and so too does the cost. The smaller the radius of curvature of the contact point on the sample, the greater the deformation or secretion required to increase the contact area.
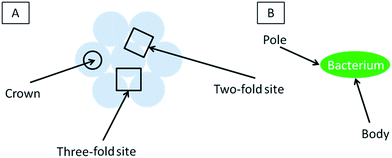
A sphere has the same curvature at every point, so there is no advantage to adsorbing on a particular point of the sphere. However, in addition to adhering to a single contact, a bacterium can increase the total area of contact by finding a second sphere to contact (providing a new contact is in range, growing a new contact requires less energy than extending an existing contact because less deformation is required). Selectivity on a uniform packed layer of spheres must arise from differences in the number of contacts that are available to a bacterium. Thus, following our assumption, a favorable position is one where the bacterium can contact several spheres, and thus where the spheres are closer together than a dimension of the bacteria. The 2-fold and 3-fold sites provide such a location whereas the crown is further away from other spheres and should be less favorable.
We now discuss these concepts of selective adsorption in relation to the experimental results. For diameters greater than 450 nm, there were no bacteria adsorbed to the crown (see Fig. 3): using the concepts above, this is because crown is the point where it is the maximum distance to another sphere for forming a second contact. On spheres in the diameter range 630–925 nm, a single bacterium is more likely to be straddling two 3-fold sites: this is because the bacterium can contact four spheres in this position. For the 230 nm particles, the discrete nature of adhesion sites is smeared out because the length and width of each bacterium is greater than the diameter of the particles: each bacterium can bind to several spheres regardless of where it sits on the surface. Because all positions on the 230 nm surface are now similar, the arrangement of the bacteria is less influenced by the topography.
It is also possible that variation in shear rate plays a role in selective occupancy of different positions. The crown projects further into the flow field than the more sheltered sites between the spheres and therefore is likely subjected to greater shear stresses that could remove bacteria.
4.2. Reduced density of bacteria on colloidal crystal vs. flat plate
The flat plate does not curve away from the bacterium, so less deformation is required to increase the contact area. Thus the flat plate is immediately more favorable for adhesion if the only consideration is the deformation required to achieve a suitable contact area.4.3. Trends in relative density of bacteria
As the radius of the particles increases, we believe that the following factors may be important: (1) less deformation is required for the same contact area on a single sphere; (2) there is an increase in the spacing between centers of the spheres, which causes a lower density of (favorable) 2-fold and 3-fold sites, (3) changes in compatibility between bacterial dimensions and binding dimensions (epitaxy) and (4) the binding sites become deeper, leading to better shelter from shear forces. The near constancy of the relative adhesion with diameter in the range 450–1550 suggests that exact matches in dimensions are not resolved or not important for the adhesion of P. aeruginosa. The first factor is expected to cause an increase in adhesion density with increasing radius whereas the second factor (diminishing density of 2-fold and 3-fold sites) should lead to an overall decrease in density of bacteria. This can also be considered as an increase in the area covered by the unfavorable crown area. The combination of opposite trends may be responsible for the fact that we observed approximately constant adhesion density as a function of diameter (Fig. 4).For 230 nm spheres, the relative density of adhesion is about the same as on the flat substrate, and significantly greater than on the larger spheres. The local curvature of each spherical particle is greater on the 230 nm particles, which means that greater deformation is required for the same area of contact at each contact – an unfavorable factor. However, the large size of P. aeruginosa (∼500 nm × ∼1300 nm) compared to the sphere diameter enables the bacterium to make contact with multiple particles (e.g. 10–20) and therefore the total bacterium-sphere area can be still high enough to favor adhesion.
4.4. Formation of colonies
In our experiments there is a correlation between selective adhesion and inhibition of colony formation. There are two possibilities that might explain this correlation. First, the locations where single bacteria are observed (2-fold or 3-fold sites) may not be conducive for biological phenomena such as reproduction or recruitment, which may inhibit colony formation. Future tests would be required to examine this possibility.Second, colony formation may be inhibited because the lack of adjacent favorable sites. The essence of a colony is close proximity (usually contact) of bacteria. But if there are no adjacent favorable sites, then some bacteria in such a colony must sit on an unfavorable site. Occupation of such a site may be so unfavorable that bacteria do not reside there and thus colonies cannot form. Referring to our colloidal crystals, for particle diameters larger than 450 nm, the individual bacteria prefer to site in the 2-fold and 3-fold sites and are not found on the crowns. To cover the entire surface would require some bacteria to sit on the less-favorable sites, and that may simply be too unattractive. Thus, one hypothesis for the lack of colony formation on particles greater than 230 nm is that adjacent favorable sites are simply too far apart.
5. Conclusions
We compared interactions of P. aeruginosa with both flat surfaces of polystyrene and surfaces coated with a monolayer of close-packed spheres. The colloidal crystal-coating reduces the number density of P. aeruginosa adhered to the surface, reduces the number of microbial colonies, and alters the arrangement of the cells. In particular: (1) the colloidal crystal reduced the number density of bacteria adhered to the surface by about 80% for particle diameters in the range 450–1550 nm; (2) spheres with diameters in the range 220–1550 nm reduced the incidence of colony formation, and no colonies were observed when the particle diameter was in the range 630–1550 nm and (3) for particle diameters exceeding 630 nm, P. aeruginosa selectively adhere in the 2-fold sites and are rarely found on the particle crown. Our results are consistent with the idea that it is more difficult for bacteria to adsorb to curved surfaces, and that bacteria select between different surface sites based on the local geometry. Furthermore, the results are consistent with the idea that the formation of colonies can be influenced by the proximity of favorable adsorption sites for individual bacteria.The incidence of bacterial colony formation on a medical grade stainless steel needle (diameter = 1.5 mm) was reduced by coating it with 450 nm particles. This result suggests that application of a layer of colloidal particles is a promising strategy to delay adhesion and colony formation of microbes on a variety of materials and geometries.
Acknowledgements
We acknowledge financial support from the David W. Francis and Lillian Francis Scholarship Fund. We also thank Dr Chang Lu, Steve Melville and Peter J. Vikesland for providing access to their laboratories at Virginia Tech. We also thank Jonathan Hittel, Akhil Jindal, Timothy Song and Nattasha Srikongyos for laboratory assistance.Notes and references
- R. M. Klevens, J. R. Edwards, C. L. Richards, T. C. Horan, R. P. Gaynes, D. A. Pollock and D. M. Cardo, Public Health Rep., 2007, 122, 160–166 Search PubMed.
- J. D. Bryers, Biotechnol. Bioeng., 2008, 100, 1–18 CrossRef CAS PubMed.
- A. Geddes, J. Antimicrob. Chemother., 2000, 46, 873–877 CrossRef CAS PubMed.
- P. S. Stewart and J. W. Costerton, Lancet, 2001, 358, 135–138 CrossRef CAS.
- F. FitzGerald, J. O'Gorman, M. M. Morris-Downes, R. K. Crowley, S. Donlon, R. Bajwa, E. G. Smyth, F. Fitzpatrick, P. J. Conlon and H. Humphreys, Journal of Hospital Infection, 2011, 79, 218–221 CrossRef PubMed.
- M. Blaser, Nature, 2011, 476, 393–394 CrossRef CAS PubMed.
- I. Banerjee, R. C. Pangule and R. S. Kane, Adv. Mater., 2011, 23, 690–718 CrossRef CAS PubMed.
- A. Marmur, Biofouling, 2006, 22, 107–115 CrossRef CAS PubMed.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Appl. Environ. Microbiol., 2013, 79, 2703–2712 CrossRef CAS PubMed.
- P. Ball, Nature, 1999, 400, 507–509 CrossRef CAS PubMed.
- C. Baum, W. Meyer, R. Stelzer, L. G. Fleischer and D. Siebers, Mar. Biol., 2002, 140, 653–657 CrossRef PubMed.
- A. Scardino, R. De Nys, O. Ison, W. O'Connor and P. Steinberg, Biofouling, 2003, 19, 221–230 CrossRef PubMed.
- K. K. Chung, J. F. Schumacher, E. M. Sampson, R. A. Burne, P. J. Antonelli and A. B. Brennana, Biointerphases, 2007, 2, 89–94 CrossRef CAS PubMed.
- K. Anselme, P. Davidson, A. M. Popa, M. Giazzon, M. Liley and L. Ploux, Acta Biomater., 2010, 6, 3824–3846 CrossRef CAS PubMed.
- C. Diaz, P. L. Schilardi, R. C. Salvarezza and M. F. L. de Mele, Langmuir, 2007, 23, 11206–11210 CrossRef CAS PubMed.
- A. K. Epstein, A. I. Hochbaum, P. Kim and J. Aizenberg, Nanotechnology, 2011, 22, 494007 CrossRef CAS PubMed.
- M. Kargar, J. Wang, A. S. Nain and B. Behkam, Soft Matter, 2012, 8, 10254–10259 RSC.
- E. Medilanski, K. Kaufmann, L. Y. Wick, O. Wanner and H. Harms, Biofouling, 2002, 18, 193–203 CrossRef.
- K. A. Whitehead, J. Colligon and J. Verran, Colloids Surf., B, 2005, 41, 129–138 CrossRef CAS PubMed.
- L. C. Xu and C. A. Siedlecki, Acta Biomater., 2012, 8, 72–81 CrossRef CAS PubMed.
- S. Y. Hou, H. A. Gu, C. Smith and D. C. Ren, Langmuir, 2011, 27, 2686–2691 CrossRef CAS PubMed.
- A. I. Hochbaum and J. Aizenberg, Nano Lett., 2010, 10, 3717–3721 CrossRef CAS PubMed.
- M. Retsch, Z. C. Zhou, S. Rivera, M. Kappl, X. S. Zhao, U. Jonas and Q. Li, Macromol. Chem. Phys., 2009, 210, 230–241 CrossRef CAS.
- J. T. Zhang, L. L. Wang, D. N. Lamont, S. S. Velankar and S. A. Asher, Angew. Chem., Int. Ed., 2012, 51, 6117–6120 CrossRef CAS PubMed.
- N. Vogel, S. Goerres, K. Landfester and C. K. Weiss, Macromol. Chem. Phys., 2011, 212, 1719–1734 CrossRef CAS.
- A. M. Brozell, M. A. Muha, B. Sanii and A. N. Parikh, J. Am. Chem. Soc., 2006, 128, 62–63 CrossRef CAS PubMed.
- S. L. Gellatly and R. E. W. Hancock, Pathog. Dis., 2013, 67, 159–173 CrossRef CAS PubMed.
- Y. H. Wang and W. D. Zhou, J. Nanosci. Nanotechnol., 2010, 10, 1563–1583 CrossRef CAS PubMed.
- Z. C. Lu and M. Zhou, J. Colloid Interface Sci., 2011, 361, 429–435 CrossRef CAS PubMed.
- R. J. Hunter, Foundations of Colloid Science, Oxford University Press, Oxford, 1987 Search PubMed.
- J. Chandra, P. K. Mukherjee and M. A. Ghannoum, Nat. Protoc., 2008, 3, 1909–1924 CrossRef CAS PubMed.
- D. M. Goeres, L. R. Loetterle, M. A. Hamilton, R. Murga, D. W. Kirby and R. M. Donlan, Microbiology-Sgm, 2005, 151, 757–762 CrossRef CAS PubMed.
- K. Buckingham-Meyer, D. M. Goeres and M. A. Hamilton, J. Microbiol. Methods, 2007, 70, 236–244 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS.
- C. Gomez-Suarez, H. J. Busscher and H. C. van der Mei, Appl. Environ. Microbiol., 2001, 67, 2531–2537 CrossRef CAS PubMed.
- P. Singleton and D. Sainsbury, in Dictionary of Microbiology and Molecular Biology, John Wiley & Sons, Ltd, 2007, pp. 451–508 Search PubMed.
- M. Kargar, A. Kim, A. S. Nain and B. Behkam, in Annual Meeting of Adhesion Society, Daytona Beach, FL, USA, 2013 Search PubMed.
- J. F. Schumacher, M. L. Carman, T. G. Estes, A. W. Feinberg, L. H. Wilson, M. E. Callow, J. A. Callow, J. A. Finlay and A. B. Brennan, Biofouling, 2007, 23, 55–62 CrossRef CAS PubMed.
- M. E. Callow, A. R. Jennings, A. B. Brennan, C. E. Seegert, A. Gibson, L. Wilson, A. Feinberg, R. Baney and J. A. Callow, Biofouling, 2002, 18, 237–245 CrossRef.
- A. J. Scardino, J. Guenther and R. de Nys, Biofouling, 2008, 24, 45–53 CrossRef CAS PubMed.
- A. J. Scardino, E. Harvey and R. De Nys, Biofouling, 2006, 22, 55–60 CrossRef CAS PubMed.
- J. F. Schumacher, C. J. Long, M. E. Callow, J. A. Finlay, J. A. Callow and A. B. Brennan, Langmuir, 2008, 24, 4931–4937 CrossRef CAS PubMed.
- K. Hori and S. Matsumoto, Biochem. Eng. J., 2010, 48, 424–434 CrossRef CAS PubMed.
- C. M. Waters and B. L. Bassler, in Annual Review of Cell and Developmental Biology, Annual Reviews, Palo Alto, 2005, pp. 319–346 Search PubMed.
- U. Seifert and R. Lipowsky, Phys. Rev. A, 1990, 42, 4768–4771 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: The results of the chemical characterization of particles and flat surface of polystyrene. Also, several optical and fluorescent images of the cells attached to the surfaces is included in order to show that SEM sample preparation dose not affect our results. See DOI: 10.1039/c4tb00835a |
| This journal is © The Royal Society of Chemistry 2014 |