Open Access Article
Open Access ArticleNanocapsules of a cationic polyelectrolyte and nucleic acid for efficient cellular uptake and gene transfer
J.
Ruesing
a,
O.
Rotan
a,
C.
Gross-Heitfeld
b,
C.
Mayer
b and
M.
Epple
*a
aInorganic Chemistry and Centre for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, 45117 Essen, Germany. E-mail: matthias.epple@uni-due.de
bPhysical Chemistry and Centre for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, 45117 Essen, Germany
First published on 20th May 2014
Abstract
Polyelectrolyte nanocapsules, consisting of poly(allylamine hydrochloride) (PAH) and a nucleic acid, i.e. either DNA or siRNA, were prepared with calcium phosphate nanoparticles as template. This inorganic core was removed by a combination of acid treatment and dialysis, leading to capsules with a diameter of about 140 nm. These capsules were well taken up by HeLa cells and led to an efficient gene transfer, i.e. transfection by DNA and gene silencing by siRNA. They behaved clearly different from unstructured aggregates of DNA and PAH, i.e. polyplexes, underscoring the effect of their internal structure.
Introduction
The layer-by-layer process where polyelectrolytes of alternating charge are assembled around a solid core1 has been successfully used to prepare microcapsules. The solid core is subsequently dissolved, leaving behind capsules which may be used to transport drugs or other biomolecules.2–11 They may also be applied as intracellular sensors, e.g. to monitor the local pH inside cells.12,13 Note that compared to microcapsules, it is more difficult to prepare nanocapsules because the separation after each coating step requires ultracentrifugation after which a redispersion is difficult.14 Besides being suitable vehicles to transport drugs and also potential reaction containers, they can be used as bioactive moieties themselves, with the biomolecules incorporated into the capsule wall. Especially interesting is the case of nucleic acids which, as polyanionic molecules, may be used for such layer-by-layer syntheses.15,16 Thus, a genetically active agent is obtained.Microcapsules of DNA and PAH were reported by Vinogradova et al.17 They used melamine–formaldehyde particles with a diameter of 4 μm as templates for the capsule preparation. Johnston and Caruso have studied oligonucleotide-based capsules.11 Wang et al. reported the synthesis of DNA/PLL capsules based on calcium carbonate cores with diameters between 1 and 4 μm.18,19 Shchukin used MnCO3 as template for DNA adsorption before further layers of chondroitin sulphate and poly(L-arginine) were added and the core was finally removed.20 Zelikin et al. used a polycation-free method to deposit DNA on silica particles and covered the DNA with thiolated polymethylacrylic acid (PMA) and poly(vinylpyrrolidone).21 Klesing et al. used nanoscopic calcium phosphate rods to deposit two layers of the cationic polymer PEI and one layer of DNA. These particles showed a high transfection efficiency.22
We have prepared nanocapsules, based on a solid core of calcium phosphate and layers of PAH and nucleic acid, and studied their effect on cells. PAH was used as a less cytotoxic cationic polyelectrolyte than PEI. Our aim was to study the physicochemical properties of these systems, and also to create an improved tool for cell transfection and gene silencing without an inorganic core.
Experimental
Synthesis of calcium phosphate nanoparticles and capsules
The PAH-coated nanoparticles were prepared according to Schwiertz et al.14 by simultaneously pumping aqueous solutions of 5 mL (NH4)2HPO4 (Sigma; c = 10.8 mM), 5 mL calcium lactate (Sigma; c = 18 mM), and 5 mL poly(allylamine hydrochloride) (PAH; Sigma, M = 56 kDa; c = 2 g L−1) into 20 mL of water. The total volume of the dispersion was 35 mL. The resulting dispersion of CaP/PAH nanoparticles was dialysed for 2 days to remove non-adsorbed PAH. The particles were then coated first with nucleic acid and then with another layer of PAH (Sigma, M = 56 kDa) and DNA as described in the following.Three different kinds of nucleic acids were used. Model DNA from herring sperm from Aldrich (≤50 b.p.) was used for experiments on the preparation and mechanical stability of the capsules (M = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–30
000–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1). Plasmid DNA (pcDNA3-EGFP) (6160 base pairs, M ≈ 4.0 × 106 Da) which encodes for enhanced green fluorescent protein (EGFP) was purified from Escherichia coli using Nucleobond® endotoxin-free plasmid DNA (Macherey-Nagel, Düren, Germany). Desalted, double-stranded siRNA from invitrogen (Paisley, UK), sense, 5′-GCAAGCUGACCCU-GAAGUUCAU-3′; antisense, 5′-AUGAACUUCAGGGU-CAGCUUGC-3′, was used for gene silencing experiments (M = 14
000 g mol−1). Plasmid DNA (pcDNA3-EGFP) (6160 base pairs, M ≈ 4.0 × 106 Da) which encodes for enhanced green fluorescent protein (EGFP) was purified from Escherichia coli using Nucleobond® endotoxin-free plasmid DNA (Macherey-Nagel, Düren, Germany). Desalted, double-stranded siRNA from invitrogen (Paisley, UK), sense, 5′-GCAAGCUGACCCU-GAAGUUCAU-3′; antisense, 5′-AUGAACUUCAGGGU-CAGCUUGC-3′, was used for gene silencing experiments (M = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021.4 Da).
021.4 Da).
For the second shell, we diluted 100 μL of the CaP/PAH nanoparticle dispersion with 700 μL ultrapure water and added 50 μL of a nucleic acid solution (either DNA or siRNA; c = 1 g L−1). In preceding experiments, this amount was found to be sufficient to reverse the surface charge of the nanoparticles, i.e. the amount of free (dissolved) DNA should be minimal. For plasmid DNA, the optimum amount was 10 μL (c = 1 g L−1), probably due to the higher molecular weight than siRNA. This led to CaP/PAH/nucleic acid nanoparticles.
For the third shell, 30 μL PAH solution (2 g L−1) was added to the CaP/PAH/nucleic acid dispersion to obtain CaP/PAH/nucleic acid/PAH nanoparticles. After preceding experiments, this amount was found to be optimal for charge reversal. To prepare the empty capsules, the pH of the dispersion of CaP/PAH/nucleic acid/PAH nanoparticles was lowered to 3 with HCl. The dispersion was subjected to dialysis in a QuixSep® microdialysis capsule covered with a Nadir® dialysis tube (Roth, pore size 25–30 Å) for one day in the presence of a cation exchanger (Serva, Serdolit Red) and then for one day in the presence of an anion exchanger (Serva, Serdolit Blue). Thereby, the dissolved calcium and phosphate ions were removed from the system, and the empty capsules remained within the dialysis tube. By Energy-dispersive X-ray spectroscopy (EDX), we analysed the composition of the nanocapsules after dialysis and ensured the complete removal of calcium phosphate.
For cell uptake experiments, we used also FITC-labelled PAH (Sigma, M = 15 kDa) to form a fluorescent outer layer.
Polyplexes, i.e. unstructured aggregates of PAH and nucleic acid, were prepared as control by simply mixing the same amounts in water: 800 μL PAH (35.7 μg L−1) plus 50 μL nucleic acid (1 g L−1) plus 30 μL PAH (2 g L−1).
To estimate the particle concentration in the dispersion, the amount of hydroxyapatite was calculated. An amount of 0.054 mmol (NH4)2HPO4 (5 mL, 10.8 mmol) equates to 0.18 mmol hydroxyapatite with n(Ca5(PO4)OH = n(NH4)2HPO4)/3. This equates to a mass of 502 g mol−1 × 0.018 mmol = 9 × 10−6 kg. A typical yield for this synthesis is 68%.23 This leads to a mass of 6.12 × 10−6 kg in 35 mL. With a density of 3160 kg m−3 this equates to a volume of 1.9 × 10−9 m3. One particle has a diameter of 120 nm (Fig. 4). The volume of one particle is 4/3π × (60 nm)3 = 9 × 10−22 m3. In the dispersion (35 mL), there are 1.9 × 10−9/9 × 10−22 = 2.2 × 1012 particles, corresponding to 2.2 × 1012/35 mL = 6.3 × 1010 particles per mL.
For cellular uptake and gene silencing experiments, we diluted 100 μL of the dispersion with 700 μL of water, 50 μL of DNA/siRNA and 30 μL of PAH solution. The particle concentration was 7.2 × 106 μL−1. For the experiments we added 50 μL dispersion (3.6 × 108 particles) to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. This leads to a concentration of 14
000 cells. This leads to a concentration of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 particles per cell. The amount of DNA/siRNA in the experiments was 50 μg/880 μL × 50 μL = 2.84 μg DNA/siRNA per well, corresponding to 0.057 μg per 25
400 particles per cell. The amount of DNA/siRNA in the experiments was 50 μg/880 μL × 50 μL = 2.84 μg DNA/siRNA per well, corresponding to 0.057 μg per 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells = 0.114 ng DNA/siRNA per cell.
000 cells = 0.114 ng DNA/siRNA per cell.
For the transfection experiments we diluted 100 μL of the dispersion with 700 μL of water 10 μL of DNA and 30 μL of the PAH solution. The particle concentration was 7.5 × 106 μL−1. For the experiments, we added 50 μL dispersion (3.75 × 108 particles) to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. This leads to a concentration of 15
000 cells. This leads to a concentration of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles per cell. The amount of DNA was 10 μg/840 μL × 50 μL = 0.57 μg per well, corresponding to 0.57 μg per 25
000 particles per cell. The amount of DNA was 10 μg/840 μL × 50 μL = 0.57 μg per well, corresponding to 0.57 μg per 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells = 0.023 ng per cell.
000 cells = 0.023 ng per cell.
Characterization techniques
Scanning electron microscopy (SEM) was carried out with an ESEM Quanta 400 FEG instrument (FEI, gold–palladium [80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20]-sputtered samples), equipped with an energy-dispersive X-ray spectrometer (Genesis 4000, SUTW-Si(Li) detector).
20]-sputtered samples), equipped with an energy-dispersive X-ray spectrometer (Genesis 4000, SUTW-Si(Li) detector).
Dynamic light scattering (DLS) and zeta potential determinations were carried out with a Zetasizer nanoseries instrument (Malvern Nano-ZS, laser: λ = 633 nm).
Atomic force microscopy (AFM) was performed with a Nanowizard AFM standard version from JPK. The diluted dispersions were dried at room temperature on a glass microscope slide. Intermittent contact mode was used with an NCH tip from Nanoworld (silicon tip with a radius of 8 nm, spring constant 42 N m−1, resonance frequency 320 kHz, scan rate 1.0 Hz).
Indentation measurements were carried in the force spectroscopy mode. The tip was horizontally positioned over the centre of the capsule and moved down 400 nm with 80 nm s−1 and then returned at the same rate to its origin position. After each indentation, the deformed capsule was again visualized in order to study the induced structural damage.
Cell experiments
For cell uptake studies, HeLa cells (human epithelial cervical cancer cell line) were cultivated in DMEM, supplemented with 10% fetal calf serum (FCS) and 100 U mL−1 penicillin/streptomycin at 37 °C under 5% CO2 atmosphere, according to standard cell culture protocols. Approximately 24 h before the experiment, the cells were trypsinized and seeded in 24-well plates with a density of 5 × 104 cells per well in 0.5 mL of cell medium.The incubation with CaP/PAH/DNA/PAH-FITC nanoparticles, the corresponding capsules and DNA/PAH-FITC polyplexes was carried out as follows. A 50 μL aliquot of each dispersion was added to the well, which corresponded to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 dilution. The cells were incubated for 3 h, then the cell culture medium was removed. The cells were washed three times with phosphate-buffered saline (PBS). The uptake of the nanoparticles, capsules or polyplexes was determined by fluorescence and transmission light microscopy (Zeiss Axiovert 40 CFL).
11 dilution. The cells were incubated for 3 h, then the cell culture medium was removed. The cells were washed three times with phosphate-buffered saline (PBS). The uptake of the nanoparticles, capsules or polyplexes was determined by fluorescence and transmission light microscopy (Zeiss Axiovert 40 CFL).
For gene silencing experiments, a HeLa–EGFP cell line was used where the cells showed a stable expression of EGFP. The cells were incubated and seeded as described above, except that the number of the cells was 2.5 × 104 per well. The cells were incubated together with nanoparticles, capsules or polyplexes for 7 h. The amount of siRNA was 2.84 μg per well. The cell culture medium was removed and fresh medium (0.5 mL) was added to each well. After 48 h of further incubation, the efficiency of gene silencing was computed as follows:
| [(Percentage of not fluorescing cells after transfection) − (percentage of not fluorescing cells in the control)]/(percentage of fluorescing cells in the control) × 100% |
HeLa–EGFP cells cultured in cell culture medium with no additives were used as control. At least 100 cells were counted in each case.
For transfection experiments, the HeLa cells were incubated and seeded as described above for gene silencing experiments, except that the incubation time after cell medium change was extended to 72 h. The amount of DNA was 0.57 μg per well. The transfection efficiency of the nanoparticles, capsules or polyplexes was determined by fluorescence and transmission light microscopy.
The transfection efficiency was calculated by the ratio of the cells in which EGFP was expressed (green fluorescence) to the total number of cells.
The cell viability was determined by the MTT assay according to the standard protocol for transfection and gene silencing experiments. Briefly, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma, Taufkirchen, Germany) was dissolved in PBS (5 mg mL−1) and then diluted to 1 mg mL−1 in the cell culture medium. HeLa cells were seeded and incubated as described above for transfection or gene silencing experiments. After 48 and 72 h, the cell culture medium of the treated cells was replaced by 300 μL of the MTT solution and incubated for 1 h at 37 °C under 5% CO2 in humidified atmosphere. Then the MTT solution was removed from the wells and 300 μL of DMSO were added to the cells. After 30 min, 100 μL aliquots were taken for spectrophotometric analysis with a Multiscan FC (Thermo Fisher scientific, Vantaa, Finland) at λ = 570 nm. The absorption of treated cells was normalized to that of control (untreated) cells, thereby indicating the relative level of cell viability.
For statistical analysis of the data, we used the ANOVA test (Analysis of Variance). p-values below 0.05 were considered as significant.
Results and discussion
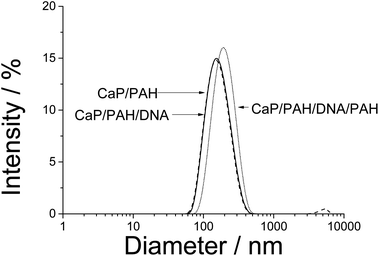
We used nanoparticles of calcium phosphate as templates for the synthesis of hollow capsules, consisting of three layers: PAH, nucleic acid, and again PAH. The capsules are held together by the electrostatic interaction between the polyelectrolyte chains. The presence of alternating layers can be demonstrated by monitoring the ζ-potential which is positive when PAH is on the outside and negative when DNA is on the outside (Fig. 1). The particle size of the calcium phosphate nanoparticles increased only moderately from 160 to 190 nm, indicating dense polyelectrolyte layers on the particle surface (Fig. 2). The polydispersity index for all samples was between 0.15 and 0.18, indicating a good monodispersity of the particles.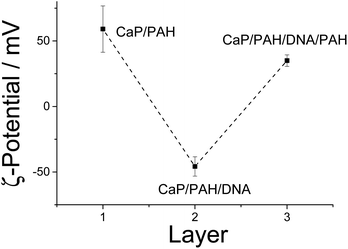 | ||
| Fig. 1 Alternating ζ-potential of calcium phosphate nanoparticles which were subsequently coated with PAH (cationic) and DNA (anionic), determined by dynamic light scattering. | ||
It was possible to remove the calcium phosphate core by dissolution in acid and absorption of the released calcium and phosphate ions by solid ion exchangers. The resulting capsules had a positive ζ-potential (+35 mV) and were only slightly smaller than the original particles (140 nm; Fig. 3).
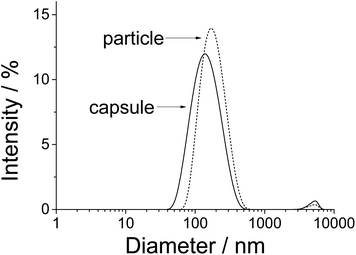 | ||
| Fig. 3 Size distribution of CaP/PAH/DNA/PAH nanoparticles and of PAH/DNA/PAH nanocapsules by dynamic light scattering. The data are representative for the three different kinds of nucleic acids. | ||
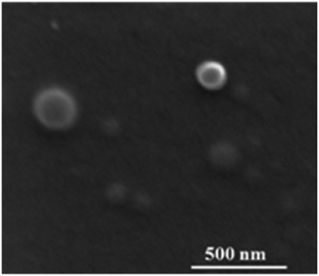
In the scanning electron microscope, the capsules were well visible as spherical objects with a diameter of 120 ± 40 nm (Fig. 4). However, it is likely that some shrinking had occurred during the drying process and in the high vacuum in the SEM. The EDX spectrum showed no signals for calcium and phosphorus (Fig. 5), confirming the quantitative removal of the calcium phosphate core.
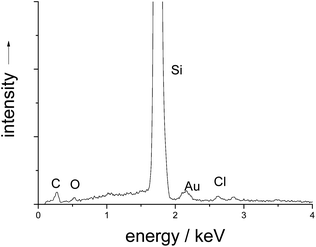 | ||
| Fig. 5 EDX spectrum of PAH/DNA/PAH nanocapsules on a silicon sample holder. Calcium and phosphorus have been quantitatively removed. | ||
Atomic force microscopy (AFM) is better suited to study such “moist” objects. Tapping-mode AFM showed spherical particles before and after removing the calcium phosphate. Nanoindentation in the AFM showed compact CaP/PAH/DNA/PAH particles (Fig. 6) and PAH/DNA/PAH capsules (Fig. 7), confirming the hollow nature of the capsules.
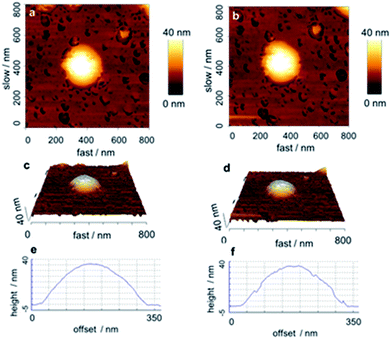 | ||
| Fig. 6 AFM-image of a CaP/PAH/DNA/PAH nanoparticle before (left) and after (right) indentation, indicating its compact and solid nature. | ||
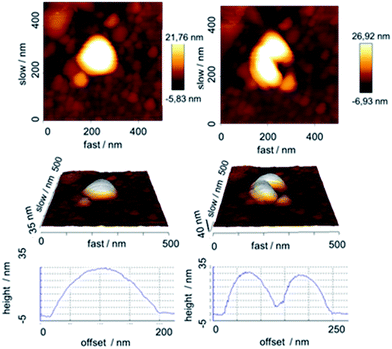 | ||
| Fig. 7 AFM-image of an air-dried PAH/DNA/PAH nanocapsule before (left) and after (right) indentation, indicating its soft and hollow nature. | ||
For comparison, we have also studied PAH/DNA polyplexes with the same ratio of PAH and DNA by dynamic light scattering. Such complexes are established in cell biology as easy-to-handle gene delivery agents (especially polyethyleneimine; PEI)24,25 despite concerns about their cytotoxicity.26 In the DLS study, the polyplexes were a highly polydisperse system of cationic aggregates with no defined structure as in the case of the capsules. Thus, we conclude that the capsules do have an internal structure that is different from an unstructured assembly of the polyelectrolytes. Fig. 8 depicts the architecture of the nanocapsules with and without solid core and also of the polyplexes.
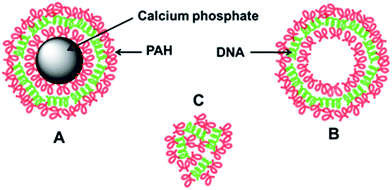 | ||
| Fig. 8 Schematic representation of the capsules with (A) and without (B) calcium phosphate core and of unstructured PAH/DNA polyplexes (C). | ||
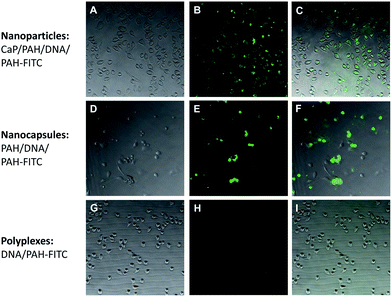
The nanoparticles, the capsules and the polyplexes were all tested with cells to elucidate whether they are taken up by cells. To this end, they were fluorescently labelled with PAH-FITC which gives a green fluorescence. Both nanoparticles and capsules were easily taken up by the cells, in contrast to polyplexes which showed a more diffuse distribution (Fig. 9).
The gene silencing efficiency was tested with EGFP-expressing HeLa cells. In this case, particles and capsules showed the efficiency of more than 75% and polyplexes up to 50% (Fig. 10).
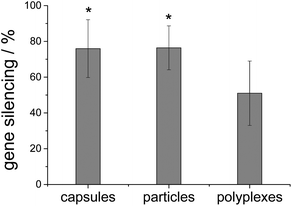 | ||
| Fig. 10 Gene silencing efficiency with siRNA on HeLa–EGFP cells. * indicates p < 0.05 in comparison to polyplexes (n = 9). | ||
The cell viability was tested by the MTT test. Particles, capsules and polyplexes showed no cytotoxicity, compared to untreated cells (100% viability) (Fig. 11).
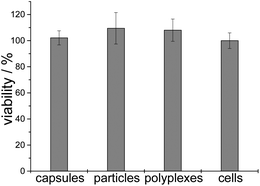 | ||
| Fig. 11 Cell viability after gene silencing experiments with siRNA (n = 6). “Cells” refers to untreated cells. | ||
The transfection experiments were carried out on HeLa cells. Note that there was no difference in transfection efficiency between particles and capsules. The transfection efficiency of PAH/DNA polyplexes was significantly lower (Fig. 12). Cell viability in all cases was near 100% related to untreated cells, which indicates no toxic effects after transfection with such agents (Fig. 13).
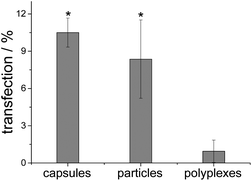 | ||
| Fig. 12 Transfection efficiency with cEGFP-DNA of nanoparticles, capsules and polyplexes. * indicates p < 0.001 in comparison to polyplexes (n = 6). | ||
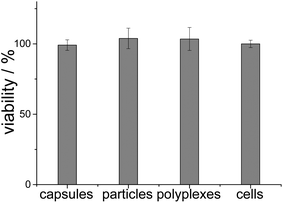 | ||
| Fig. 13 Cell viability after transfection experiments with cEGFP DNA (n = 6). “Cells” refers to untreated cells. | ||
In summary, we have shown that coated calcium phosphate nanoparticles and the nanocapsules that remain after the removal of the inorganic core have the same transfection and gene silencing efficiency. This demonstrates that a “hard core” is not necessary for cellular uptake and that nanocapsules can serve as well-defined delivery systems into cells. Kastl et al. have recently studied the cellular uptake of microcapsules in detail and followed their intracellular localization.27 This indicates a different biological effect compared to nanoparticles and polyplexes and may explain the different performance, compared to unstructured polyplexes. Mechanistically, we assume that capsules and particles are both taken up by endocytosis, ending up first in an endosome and then in a lysosome where the proton-sponge effect of PEI and calcium phosphate leads to lysosomal escape and release of the nucleic acids into the cytoplasm.28
Conclusions
Calcium phosphate nanoparticles were successfully functionalized by two layers of PAH and one layer of DNA or siRNA and used for transfection and gene silencing. The calcium phosphate core was removed with HCl. Particles and capsules showed similar transfection efficiencies and gene silencing efficiencies. Thus, the internal structure of a nanocapsule controls their uptake mechanism and clearly distinguishes it from an unstructured polyplex of DNA and cationic polyelectrolyte.Notes and references
- G. Decher, Science, 1997, 277, 1232–1237 CrossRef CAS.
- W. Tong, X. Song and C. Gao, Chem. Soc. Rev., 2012, 41, 6103–6124 RSC.
- B. G. de Geest, G. B. Sukhorukov and H. Mohwald, Expert Opin. Drug Delivery, 2009, 6, 613–624 CrossRef CAS PubMed.
- W. Tong and C. Gao, J. Mater. Chem., 2008, 18, 3799–3812 RSC.
- C. A. Jewell and D. M. Lynn, Adv. Drug Delivery Rev., 2008, 60, 979–999 CrossRef CAS PubMed.
- S. A. Sukhishvili, Curr. Opin. Colloid Interface Sci., 2005, 10, 37–44 CrossRef CAS PubMed.
- C. Mayer, Int. J. Artif. Organs, 2005, 28, 1163–1171 CAS.
- D. G. Shchukin and G. B. Sukhorukov, Adv. Mater., 2004, 16, 671–682 CrossRef CAS.
- C. S. Peyratout and L. Daehne, Angew. Chem., Int. Ed., 2004, 43, 3762–3783 CrossRef CAS PubMed.
- R. de Rose, A. N. Zelikin, A. P. R. Johnston, A. Sexton, S. F. Chong, C. Cortez, W. Mulholland, F. Caruso and S. J. Kent, Adv. Mater., 2008, 20, 4698–4703 CrossRef CAS.
- A. P. R. Johnston and F. Caruso, Angew. Chem., 2007, 119, 2731–2734 CrossRef.
- P. R. Gil, M. Nazarenus, S. Ashraf and W. J. Parak, Small, 2012, 8, 943–948 CrossRef PubMed.
- L. L. Mercato, A. Z. Abbasi and W. J. Parak, Small, 2011, 7, 351–363 CrossRef PubMed.
- J. Schwiertz, W. Meyer-Zaika, L. Ruiz-Gonzalez, J. M. Gonzales-Calbet, M. Vallet-Regi and M. Epple, J. Mater. Chem., 2008, 18, 3831–3834 RSC.
- X. Zhang, A. Kovtun, C. Mendoza-Palomares, M. Oulad-Abdelghani, S. Facca, F. Fioretti, J. C. Voegel, M. Epple and N. Benkirane-Jessel, Biomaterials, 2010, 31, 6013–6018 CrossRef CAS PubMed.
- T. Boudou, T. Crouzier, K. Ren, G. Blin and C. Picart, Adv. Mater., 2010, 22, 441–467 CrossRef CAS PubMed.
- O. I. Vinogradova, O. V. Lebedeva, K. Vasilev, H. Gong, J. Garcia-Turiel and B. S. Kim, Biomacromolecules, 2005, 6, 1495–1502 CrossRef CAS PubMed.
- Z. Wang, L. Qian, X. Wang, H. Zhu, F. Yang and X. Yang, Colloids Surf., A, 2009, 332, 164–171 CrossRef CAS PubMed.
- Z. Wang, L. Qian, X. Wang, F. Yang and X. Yang, Colloids Surf., A, 2008, 326, 29–36 CrossRef CAS PubMed.
- D. G. Shchukin, A. A. Patel, G. B. Sukhorukov and Y. M. Lvov, J. Am. Chem. Soc., 2004, 126, 3374–3375 CrossRef CAS PubMed.
- A. N. Zelikin, A. L. Becker, A. P. R. Johnston, K. L. Wark, F. Turatti and F. Caruso, ACS Nano, 2007, 1, 63–69 CrossRef CAS PubMed.
- J. Klesing, S. Chernousova and M. Epple, J. Mater. Chem., 2012, 22, 199–204 RSC.
- V. Sokolova, T. Knuschke, J. Buer, A. M. Westendorf and M. Epple, Acta Biomater., 2011, 7, 4029–4036 CrossRef CAS PubMed.
- A. Akinc, M. Thomas, A. M. Klibanov and R. S. Langer, J. Gene Med., 2005, 7, 657–663 CrossRef CAS PubMed.
- O. Boussif, F. Lezoualch, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J. P. Behr, Proc. Calif. Acad. Sci., 1995, 92, 7297–7301 CAS.
- L. Parhamifar, A. K. Larsen, A. C. Hunter, T. L. Andresen and S. M. Moghimi, Soft Matter, 2010, 6, 4001–4009 RSC.
- L. Kastl, D. Sasse, V. Wulf, R. Hartmann, J. Mircheski, C. Ranke, S. Carregal-Romero, J. A. Martínez-López, R. Fernández-Chacón, W. J. Parak, H. P. Elsasser and P. Rivera-Gil, ACS Nano, 2013, 7, 6605–6618 CrossRef CAS PubMed.
- V. Sokolova, D. Kozlova, T. Knuschke, J. Buer, A. M. Westendorf and M. Epple, Acta Biomater., 2013, 9, 7527–7535 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |