Open Access Article
Open Access ArticleMembrane curvature based lipid sorting using a nanoparticle patterned substrate†
Joshua C.
Black
,
Philip P.
Cheney
,
Travis
Campbell
and
Michelle K.
Knowles
*
Department of Chemistry and Biochemistry, University of Denver, Denver, CO 80208, USA. E-mail: michelle.knowles@du.edu
First published on 15th January 2014
Abstract
Cellular membranes contain a variety of shapes that likely act as motifs for sorting lipids and proteins. To understand the sorting that takes place within cells, a continuous, fluid bilayer with regions of membrane curvature was designed and characterized using confocal fluorescence and total internal reflection fluorescence microscopy techniques. A supported lipid bilayer was formed over fluorescently labelled nanoparticles deposited on a glass surface. The lipid composition and membrane shape are separately controlled and the nanoparticle dimensions (d = 40–200 nm) determine the extent of curvature. The bulk membrane is fluid as demonstrated by fluorescence recovery after photobleaching (FRAP) using dye labelled lipids. In bilayers that contain fluorescently labelled, single-tailed lipids, accumulation is observed at regions of curvature, yet the molecules retain fluidity. Using single particle imaging methods, lipids are observed to visit regions of curvature and exchange with the surrounding flat membrane. The nanoparticle patterned substrate described here allows for quantitative measurement of the transient interactions between fluorescently labelled biomolecules and regions of membrane curvature.
Introduction
The shape of lipid membranes plays a fundamental role in biological systems by facilitating protein and lipid sorting.1–5 The presence of curvature in a membrane is predicted to stabilize lipid microdomains6,7 and has been shown to alter protein function.8,9 Lipid composition varies throughout membrane trafficking pathways as does membrane curvature, suggesting a role for curvature as a means of sorting lipids. In living cells, the variation of lipid tails is enough to allow for differentiated distribution of lipids.10 However, both local changes to the lipid composition and recruited proteins can alter membrane curvature,11 creating interplay between the shape of a membrane and the chemical composition.Proteins that sense or generate membrane curvature have been well characterized and often include amphipathic helices or Bin/Amphiphysin/Rvs (BAR) domains.11,12 Three main mechanisms of protein binding to regions of membrane curvature have been reported. First, amphipathic helices are proposed to insert into hydrophobic regions.13,14 It is thought that more hydrophobic “defect” sites are accessible when a lipid membrane is formed into a highly curved (d < 100 nm) membrane. Second, other proteins, such as those that contain BAR domains, have an intrinsic curvature that allows them to bind with higher affinity when the membrane curvature matches well to the shape of the protein.15,16 Lastly, some endocytic proteins, such as clathrin, build scaffolds and those larger structures have a curvature that interacts with lipid membranes, often conforming membranes to match the scaffold shape.17
Likewise, several mechanisms are proposed for lipid sorting at regions of membrane curvature. One mechanism implies that lipids bind to the hydrophobic defect sites that are exposed on regions of curvature,14,18 similar to the mechanism employed by amphipathic helices. A second mechanism of lipid binding involves geometric constraints where the molecular shape of the lipid determines how well it fits in the curved region. For example, lipids that have an inverse conical shape, such as lysophosphatidylcholine, favor regions of positive, or convex, curvature.19 However, the shape of a molecule is not thought to be the driving force of lipid sorting in biological systems.20 The third mechanism of membrane curvature sensing involves domains of lipids as the curvature sensors. Lipid domains have unique physical properties that are not present when only a single molecule is sensing. Lipid domain properties, such as a low bending modulus, give rise to a curvature preference.21 This is supported by studies in model systems where transient lipid clusters ranging from a few nanometers to 100 nm in diameter are stabilized by fluctuations in membrane curvature.6,7 Additional factors can affect lipid sorting, such as the velocity at which a membrane is deformed22,23 or protein interactions.24
To understand how biomolecules sense curvature, in vitro biochemical systems that allow for separate tuning of curvature and lipid composition are needed. To date, liposomes,14,20 lipid coated nanoparticles in solution,3 lipid tubes,2,25 and patterned surfaces1,26,27 have provided tools for understanding the effect of lipid curvature on sorting. Each assay has limitations, many of which are overcome with the nanoparticle patterned substrate described here. For example, in liposome based assays, polydispersity in size is an issue but overcome with single liposome based assays.14,28 However, liposomes lack a continuous membrane connecting flat and curved regions and measurement of dynamics is not possible. In other assays, ensemble measurements are made.20,25,26 Finally, in assays that have connected regions of positive and negative membrane curvature, single molecule imaging has been demonstrated,1 but the high extent of curvature, meant to mimic exocytic and endocytic processes in the substrate described here, is novel. Although all of these assays have their strengths, the nanoparticle patterned substrates characterized here add a missing tool to the toolbox of biochemically based, membrane curvature assays.
Here, we have designed a nanoparticle patterned substrate that contains a continuous membrane with flat and curved regions in dynamic equilibrium and used this to assess curvature based lipid sorting. Curved, supported lipid bilayers were formed over top of fluorescent, polystyrene nanoparticles (d = 40–200 nm) deposited on a glass surface (Fig. 1A). The fluidity of the lipid bilayer was assessed with fluorescence recovery after photobleaching (FRAP) techniques. Three strengths of this nanoparticle patterned substrate are: (i) membrane curvature and lipid composition can be separately controlled. In a cellular environment, this is a challenge as curved membranes accumulate different lipids, muddling the interpretation of what drives protein or lipid sorting. (ii) Single molecule imaging is possible. We demonstrate that single fluorescent lipids can be followed in time using total internal reflection fluorescence (TIRF) microscopy. (iii) Curvatures smaller than the diffraction limit can be observed. Because the nanoparticle template determines the shape of the overlying lipid bilayer, sub-diffraction limited, fluorescent nanoparticles can be imaged alongside fluorescently labeled lipids and proteins. This allows us to mimic small vesicular structures such as a budding, clathrin coated, endocytic vesicle29 or a recently fused synaptic vesicle that has not flattened completely.30
Using nanoparticle patterned substrates we observe lipids sorting as a function of curvature. A fluorescein labeled fatty acid (5-hexadecanoylaminofluorescein) is enriched at sites of curvature, with the density of accumulation increasing with higher regions of curvature. These lipid domains are capable of exchanging with the flat membrane surface, as shown by the FRAP recovery of membrane curvature bound clusters. Lastly, the dynamics of single lipids near sites of curvature were visualized using TIRF microscopy. This is the first membrane curvature assay that contains spherically curved structures in continuum with flat membranes, allowing for characterization of the dynamics involved with lipid curvature sensing.
Experimental methods
Membrane curvature assay
Nanoparticle patterned substrates were created in 8-well dishes (Lab-Tek® Chambered Borosilicate Coverglass System, Thermo Fisher) that contain 1.5 thickness glass coverslips on bottom. Glass surfaces were cleaned extensively by soaking in SDS (0.1% w/v) for 1 hour, rinsing with DI water then soaking in a 1% v/v bleach solution overnight. Cleaned coverslips were stored in DI water until use. Immediately prior to nanoparticle sedimentation and the formation of lipid bilayers, coverslip wells were filled with 100 μL of 2% Hellmanex (Hellma, Mullheim, Germany) then rinsed with a deposition buffer (30 mM HEPES, 2 mM CaCl2, 140 mM NaCl, pH 6.4).Fluorescent, polystyrene nanoparticles were diluted in deposition buffer, sonicated, and added to the cover glass. Sedimentation occurred over the course of 30 minutes at room temperature and contact with the glass surface was sufficient for immobilization. For all measurements, deposition was optimized to keep the density of nanoparticles between 0.05 and 0.1 nanoparticle per μm2. For confocal imaging experiments, red fluorescent (580/605) polystyrene nanoparticles were used. For TIRF microscopy, yellow fluorescent (505/515) polystyrene nanoparticles were used. Nanoparticle surfaces were coated with lipids using standard liposome deposition techniques.31 Briefly, lipids films were made by drying chloroform solvated lipids under vacuum. Lipids were resuspended in deposition buffer and liposomes were created using a probe sonicator. The cover glass containing nanoparticles was incubated with liposomes at 37 °C for 1 hour. With a continuous surface, binding can occur anywhere then proceed over 1 hour of incubation at 37 °C to equilibrium. Based on our observed rate of lipid diffusion and that reported in supported lipid bilayers,32,33 1 hour is more than adequate for lateral lipid diffusion. Prior to imaging, the bilayer surface was washed three times by the incremental addition and removal of the deposition buffer to remove remaining liposomes. Lipid films contained soy phosphatidylcholine (SoyPC) and either DHPE (1,2-dihexadecanoyl-sn-glycero-3-phosphoethanol-amine) covalently bound to Marina Blue (MB-DHPE) or 5-hexadecanoylaminofluorescein (Fl-HDA). The mole ratio of SoyPC to labeled lipids was 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 unless otherwise specified.
2 unless otherwise specified.
To create asymmetrically labeled bilayers, a POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer was created then incubated with fatty acid free BSA that had been loaded with Fl-HDA for one hour following previously published work10 and separated by extensive dialysis in phosphate buffered saline PBS. All fluorescent lipids and nanoparticles were purchased from Invitrogen. Buffer components were purchased from Sigma Aldrich. SoyPC and POPC were purchased from Avanti Polar Lipids (Alabaster, AL).
Confocal fluorescence microscopy and image analysis
Images were taken using an Olympus FV1000 Laser Scanning Confocal Microscope. Nanoparticles were excited using a 594 nm diode laser; MB-DHPE, with 405 nm; and Fl-HDA with 488 nm. FRAP images were acquired at a rate of 2 μs per pixel with an image size of 512 by 512. Lipids were bleached for 0.8 seconds in a 10.3 μm diameter circle. Images of nanoparticles and fluorescein labeled fatty acids were acquired at a rate of 12.5 μs per pixel.Images were analyzed using ImageJ and MATLAB. All FRAP images were adjusted for image photobleaching by correcting the intensity to a reference area outside the bleached area. The FRAP average intensities were normalized to the highest (typically the first frame, pre-bleach) and lowest values (the bleach frame) by assigning the highest intensity to be 1.0 and the lowest to be 0.0. The mobile fraction was calculated by averaging the normalized intensity of the final three time points and dividing by the average normalized intensity of the three time points prior to bleach.
Colocalization analysis was performed by locating nanoparticles in the red images using an algorithm from the MOSAIC tracking software34 and an established colocalization routine.35,36 Around each nanoparticle location found in the red channel, a 25 × 25 pixel square was cropped from both the green channel and the red channel using a home-built MATLAB program. This stack of images was then averaged. Spot finding was not performed for the green channel. To calculate the intensity of individual spots, an annulus around the center, an estimate of the local background intensity, was subtracted from the center intensity.35 To randomize the red image, the JaCoP plug-in was used37 and 4 × 4 pixel regions were cut from data images and randomly assembled to make a new image. The cross-section of the average cropped image was normalized by dividing by the lowest intensity value. The fluorescent lipid concentration was kept constant and the background variability was minimal. To correct for the surface area differences between 40, 100, and 200 nm nanoparticles, we followed established methods.38 The nanoparticle site of curvature was approximated as a cylinder with a height equal to half of the nanoparticle diameter that has a hemisphere sitting on top.
Total internal reflection fluorescence microscopy
Single particle tracking experiments were performed using a TIRF microscope (Nikon) equipped with a 491 nm laser for excitation of the yellow-green, polystyrene nanoparticles, a 561 nm laser for excitation of Alexa Fluor 546 labeled streptavidin (Invitrogen), a 60× TIRF objective (NA 1.49), a 2.5× magnifier, and an EMCCD camera (Andor iXon 897+) for detection. The nanoparticles are stationary and an image of the nanoparticles was taken prior to dynamic measurements of the streptavidin tagged lipids. A dual-color, TIRF filter set was used (Chroma, Bellows Falls, VT) with emission filters (Omega Optical, Brattleboro, VT) at 525/45 nm and 595/60 nm. Image series were captured at 15 Hz using MicroManager.39Results and discussion
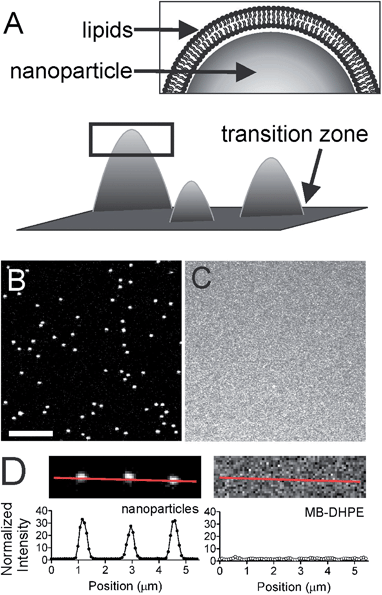
Design of a lipid bilayer with regions of high curvature
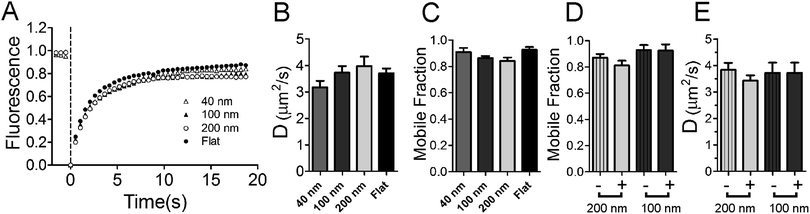
To understand the mechanism of curvature sensing, a surface that contains regions of spherically curved membranes in continuum with flat membrane surfaces was created (Fig. 1A). Nanoparticles (d = 40–200 nm) were deposited on clean glass surfaces, coated with a lipid bilayer using standard liposome deposition techniques, and imaged on a point-scanning, confocal microscope. Red-fluorescent nanoparticles mark regions of membrane curvature (Fig. 1B) and lipids are evenly distributed, as shown by the uniform fluorescence of a blue fluorescent lipid, MB-DHPE (Fig. 1C). To assay the uniformity of the samples, a fluorescence intensity line scan over the area marked by the red lines in Fig. 1D were measured. The variation in intensity of MB-DHPE is much less than the variation in fluorescence intensity of the nanoparticles. If a labeled lipid bound to sites of curvature, the fluorescent lipid would accumulate in spots containing fluorescent nanoparticles.To assess whether or not the presence of nanoparticles affected the fluidity of the lipid membrane, fluorescent lipids were photobleached and the rate of recovery was measured using standard FRAP techniques. Lipid recovery traces were measured for bilayers that were created over nanoparticles ranging from 40–200 nm in diameter (Fig. 2A). The recovery was compared to samples that contained no nanoparticles (“flat” membranes). Data was normalized and corrected for bulk photobleaching due to imaging as described in the Methods. From the recovery, the diffusion constant, D (μm2 s−1), was calculated according to established methods:40
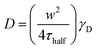 | (1) |
| F(t) = F0 + (F∞ − F0)(1 − e−kt) | (2) |
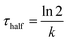 .
.
Both the rate of diffusion and the fraction mobile (Fig. 2B and C) are unaltered with the presence of nanoparticles and are in good agreement with previous FRAP studies on supported lipid bilayers.32,41 There is no significant difference observed between any groups (P-value for 〈D〉 = 0.1951, P-value for fraction mobile = 0.0605, ANOVA testing). The fluidity of lipids is maintained at nanoparticle locations (Fig. 2D and E). Small regions (570 nm diameter, circular regions) were selected at nanoparticle positions (+) and non-nanoparticle positions (−) on the supported lipid bilayer. The nanoparticles add surface area to the sample; when using 200 nm diameter nanoparticles, approximately 36% of the lipids within this small circle are on the nanoparticle and the rest are present in an annulus around the nanoparticle. With such a large percentage of lipids on the nanoparticle, if the nanoparticle binds lipid like large (d = 16 μm) polystyrene microspheres do (Fig. S1†), fluorescence recovery would be detectably hindered. We assume that lipids that bind tightly to polystyrene are completely incapable of moving, as observed in Fig. S1,† and we expect that the rate of diffusion and the fraction mobile to measurably decrease. The mobile fraction is 0.87 when no nanoparticles are present. If the nanoparticle hindered diffusion, recovery would be only (0.87 × (1 − 0.36)) = 0.56 for 200 nm nanoparticles. Lipids recover equally well when 100 nm nanoparticles are present and when they are not (Fig. 2D and E). Additionally, lipids were also observed to surround and recover on large (d = 1 μm) nanoparticles (Fig. S2†), suggesting that lipids do not interact specifically with the carboxylate modified polystyrene surface of the nanoparticles when a continuous bilayer is deposited. It is unlikely that the nanoparticle surface is greatly affecting fluidity on the nanoparticle patterned lipid bilayers.
The nanoparticles used were diffraction limited in size, making the direct imaging of lipid coverage and the transition zone between the regions of positive curvature and the flat surface difficult. To address whether or not the lipids coat the nanoparticles, a large nanoparticle (d = 1 μm) was imaged and lipids were observed to fully cover the nanoparticle (Fig. S2†). Regarding the transition zone, we expect the lipids to follow around the nanoparticles as they transition from the curved surface to the flat surface based on the work of others.42 This creates a transition zone where negative curvature exists in between the positive and flat regions that is difficult to directly assess even by AFM.43,44 The nature of this transition is not clear; lipids could form a tent-like structure (depicted in Fig. 1A) with a gentle negative curvature, or the lipids could wrap tightly to the nanoparticle, creating more of a neck-like structure as seen in fusion pores and endocytic vesicles.45,46 Our data suggest that a tent-like structure is favored based on the following argument. Lipids bilayers are typically not fluid on polystyrene (Fig. S1†), but become more fluid when the particle diameter is smaller (d = 1 μm) and connected to a flat surface (Fig. S2†). Decreasing the particle diameter further (d = 100–200 nm) leads to more fluidity when connected to the flat membrane as measured by the diffusion coefficient and the mobile fraction in FRAP experiments (Fig. 2D and E). This implies that contact between the lipids and nanoparticles is less than predicted by a tight wrapping of lipids around the nanoparticle surface. A tent-like transition zone, where many of the lipids are not in direct contact with the nanoparticle surface through the transition from positive curvature on the nanoparticle to flat surfaces, is likely.
Regions of membrane curvature recruit a single-tailed lipid
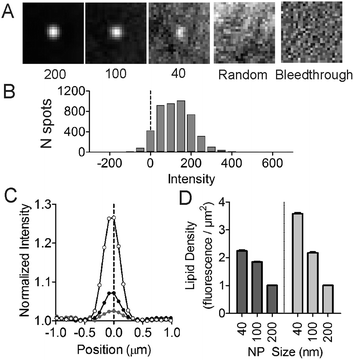
Next, we assessed the ability of lipids to sort as a function of membrane curvature. Molecules that sense curvature have increased fluorescence spatially coincident with nanoparticle locations; those that do not will have uniform fluorescence across the field of view, like that of MB-DHPE (Fig. 1). Fluorescent nanoparticles, which mark sites of membrane curvature, are imaged in one color channel, followed by imaging of fluorescently labeled lipids in a different color channel. To quantify the ability of lipids to bind to curved membranes, colocalization measurements were performed. The fluorescent intensity from the lipid channel that colocalized to nanoparticles represents the extent to which lipids bind to regions of curvature.The spatial distribution of a single-tailed lipid, fluorescein-labeled hexadecanoic acid (Fl-HDA), was evaluated on the nanoparticle patterned surface. Fl-HDA (2%) was added to lipid films and colocalizes with regions of membrane curvature (Fig. 3). Red fluorescent nanoparticles (d = 200 nm) and Fl-HDA were imaged separately and an overlay reveals that most nanoparticles accumulate Fl-HDA at the concentration used (Fig. 3A–C). To quantify the ability of regions of membrane curvature to bind lipids, a colocalization method was performed that involved finding sites of curvature using an automated algorithm. The red nanoparticles were used to find locations of curvature and the regions identified were transferred to the green image. Images were cropped to a 25 × 25 pixel (2 × 2 μm2) area around each region from both the green (Fl-HDA) and red (nanoparticle) channels.36 If a green spot appears in the center, the two differently colored species are colocalized and the extent of colocalization is represented by the intensity. The average cropped image for Fl-HDA clearly reveals preferential binding at sites of curvature for all sizes (Fig. 4A). Cropping random spots (a negative control) and measuring the green intensity when no lipids are present (a bleedthrough control) shows no spot in the center, even when contrast is enhanced (Fig. 4A).
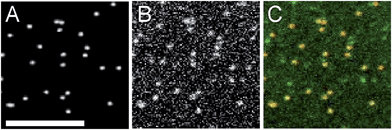 | ||
| Fig. 3 Fluorescein labeled hexadecanoic acid accumulates in regions of curvature. (A) Images of nanoparticles, (B) Fl-HDA, and (C) an overlay are shown. Scale bar = 5 μm. | ||
The intensity was quantified in two ways. First, the intensity within the center (with a local background subtracted) was calculated for each region of curvature. A histogram of the distribution of intensities observed (Fig. 4B) reveals that most nanoparticles collect single tailed lipids. Second, by plotting a line scan of the average cropped images the average accumulation is evaluated. The normalized line scan conveys the enrichment of the green fluorescent lipids for sites of curvature (Fig. 4C). Although there is significantly more green fluorescence coincident with 200 nm nanoparticles, once the surface area difference is corrected for, the density of Fl-HDA is higher on smaller nanoparticles or higher curvatures (Fig. 4D). Regions with higher curvatures recruit curvature-sensing lipids at a higher density.
To test if the recruitment of single tailed lipids depends on lipid demixing, nanoparticle patterned substrates were created with a POPC bilayer formed over top of nanoparticles instead of the SoyPC lipid mixture. There is no qualitative difference between Fl-HDA sorting using substrates containing POPC (Fig. S3†), which cannot form domains, and SoyPC (Fig. 3 and 4). Even though membrane curvature has been shown to provoke demixing of lipids in a system near a demixing point,47 and can stabilize lipid rafts48 our data suggest that SoyPC is not demixing into lipid domains at sites of membrane curvature to facilitate sorting.
We demonstrate that membrane curvature can provide a means for sorting lipids. Single tailed lipids preferentially bind to sites of curvature with higher densities at higher curvatures. This is observed in other work with single liposomes (d ∼ 50–500 nm).14 Although the curvature selectivity we observe is less than previously reported, this may be due to the symmetric labeling method used. When asymmetric bilayers are created, more selectivity is observed (Fig. 4D), however, the precise concentration of Fl-HDA incorporated into the membrane is not well-defined. The asymmetrically bilayers were prepared with Fl-HDA in the top leaflet by loading BSA with Fl-HDA and allowing it to exchange with a bilayer formed over top of nanoparticles.10 We see no qualitative difference between asymmetrically loaded bilayers (Fig. S3† and 4D) and symmetrically loaded bilayers (Fig. 3 and 4). Both show Fl-HDA accumulation at regions of membrane curvature and a higher density of Fl-HDA bound to membranes with higher curvature. However, asymmetrically labeled bilayers recruit even higher densities of Fl-HDA. This difference suggests that the observed intensity at regions of positive curvature, an “inclusive” effect, in a symmetrically labeled bilayer may be slightly less than expected due to an “exclusive” effect, where lipids are excluded from the regions of negative curvature present on the inner leaflet near the nanoparticle. Based on previous work, a defect model was proposed where regions with higher curvature had more binding sites for curvature seeking molecules.14 This model does not assume that when molecules bind to regions of positive curvature that they are excluded from regions of negative curvature with equal and opposite magnitude. The alternative geometric model suggests that the molecular shape of a lipid can give rise to curvature preference, such that inverse conical shaped lipids, like lysoPC, would prefer regions of positive curvature and avoid regions of negative curvature. Our data support a defect site model with more defect sites on higher regions of curvature.
Lipids that are enriched at sites of curvature maintain dynamic equilibrium with surroundings
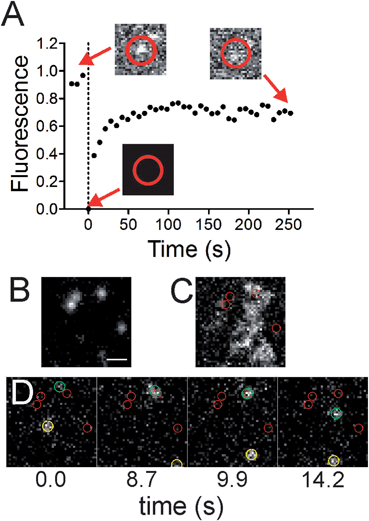
The colocalized lipids are in dynamic equilibrium with the surroundings. Photobleaching of Fl-HDA spots that reside at sites of membrane curvature (Fig. 5A) show recovery, however the rate of diffusion is approximately 40% slower at regions of curvature when compared to flat regions. To demonstrate that Fl-HDA clusters exchange with their surroundings, a single, nanoparticle and its corresponding Fl-HDA channel are imaged prior to bleaching. After a bleach to background-level signal (t = 0.0 s), the signal colocalized with the nanoparticle recovered to ∼80% after 4 minutes. The rate of recovery is significantly slower than that of MB-DHPE (Fig. 2), indicating that Fl-HDA is interacting differently than MB-DHPE when curved membranes are present. Further qualitative interpretation of the recovery of fluorescence over a nanoparticle (Fig. 5A) suggests that lipids do not migrate as a patch to recover; jumps in intensity are not observed during the recovery. Even in individual traces (data not shown) a gradual increase in fluorescence is observed. This suggests that lipids continuously appear, possibly recruited individually to regions of curvature.Direct observation of two-tailed, lipid molecules (DHPE) among regions of curvature was obtained with single molecule imaging techniques. Green fluorescent nanoparticles were imaged (Fig. 5B) to identify regions of curvature, followed by imaging of biotinylated DHPE molecules that were tagged with Alexa546-labeled streptavidin (Fig. 5C and D). These measurements were performed on 40 nm nanoparticles surfaces to avoid the challenges that the exponentially decaying, evanescent field creates. With larger nanoparticles, the incident angle would need to be adjusted to allow for visualization of lipids that are on top of nanoparticles. The dynamics of the lipids were recorded in time and the average projection of the image sequence (Fig. 5C) reveals where the lipids resided. If a lipid interacts favorably with a site of curvature and spends more time there, a brighter spot would be seen in the average projection. Although no accumulation is observed, an example series of images shows a lipid molecule visiting a curvature site and also diffusing on flat surface (Fig. 5D). Recruitment of individual lipids is not observed and previous theoretical and experimental work suggests that single lipids are not able to sense curvature.2,47
The fluorescently labeled, single-tailed lipids were enriched at sites of curvature, yet they were in dynamic equilibrium with the surrounding flat membranes. We propose two interpretations of this data. First, if we consider a defect model for lipid binding, where the density of defect sites increases at regions of positive curvature and curvature sensing molecules bind to defect sites,14,18 then the mobility observed could represent how the defect sites exchange between flat and curved substrates through the barrier of the transition zone. In this model the bending of a phospholipid bilayer to conform to regions of positive curvature induces packing defects, where hydrophobic lipid tails readily bind. A second interpretation is that defect sites are static and curvature sensing molecules bind reversibly between defect sites and non-defect sites. To move from the site of curvature, molecules need to dissociate from the defect site and laterally diffuse. It is not yet clear whether there are two types of binding that can occur between lipids and how binding to a defect site is molecularly different from other ways of being incorporated in a bilayer.
The latter model, where lipids associate and dissociate from defect sites while remaining in the lipid bilayer, suggests that more than one type of interaction can occur between lipids and regions of membrane curvature. In light of this, techniques that can measure dynamic heterogeneity will be essential. A major strength of the nanoparticle patterned substrate described here is its usefulness in single particle tracking techniques. Using such methods, the distribution of dynamics involved with membrane curvature sensing can be measured. We foresee this substrate as being highly useful when measuring transient, heterogeneous interactions between lipids or proteins and regions of membrane curvature.
Conclusions
In the current work, a nanoparticle patterned substrate that contains regions of membrane curvature is described. The extent of curvature and the lipid composition can be separately adjusted to understand the role of each in membrane curvature sensing. We have demonstrated that the curved membrane substrates are useful for both single molecule imaging experiments to characterize the dynamics of sorting, and colocalization measurements to characterize the recruitment of lipids to regions of curvature. These engineered membranes have unique topological features meant to mimic the diverse structures observed within cells and provide a new tool for characterizing membrane curvature sensing. The curved membrane substrate was used to characterize the sorting of lipids, where higher curvatures accumulated a higher density of lipids. Future studies can elucidate the mechanism by which lipids and proteins sort based on membrane shape.Acknowledgements
PPC was partially supported by the Chickasaw Nation. TC received funding from the University of Denver Undergraduate Research Center Partners in Scholarship program. This work was supported by the National Science Foundation Division of Chemical, Bioengineering, Environmental, and Transport Systems (NSF-CBET) Grant # 1033215. We thank Scott M. Reed for critical proofreading of the manuscript.References
- R. Parthasarathy, C. H. Yu and J. T. Groves, Langmuir, 2006, 22, 5095–5099 CrossRef CAS PubMed.
- A. Tian and T. Baumgart, Biophys. J., 2009, 96, 2676–2688 CrossRef CAS PubMed.
- M. S. Wang, R. E. Messersmith and S. M. Reed, Soft Matter, 2012, 8, 3909–3918 Search PubMed.
- H. Gerlach, V. Laumann, S. Martens, C. F. Becker, R. S. Goody and M. Geyer, Nat. Chem. Biol., 2010, 6, 46–53 CrossRef CAS PubMed.
- M. B. Pedersen, X. Zhou, E. K. Larsen, U. S. Sorensen, J. Kjems, J. V. Nygaard, J. R. Nyengaard, R. L. Meyer, T. Boesen and T. Vorup-Jensen, J. Immunol., 2010, 184, 1931–1945 CrossRef CAS PubMed.
- S. Meinhardt, R. L. Vink and F. Schmid, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4476–4481 CrossRef CAS PubMed.
- M. Schick, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 85, 031902 CrossRef CAS.
- J. Bigay, P. Gounon, S. Robineau and B. Antonny, Nature, 2003, 426, 563–566 CrossRef CAS PubMed.
- U. B. Jensen, E. E. Ferapontova and D. S. Sutherland, Langmuir, 2012, 28, 11106–11114 CrossRef CAS PubMed.
- S. Mukherjee, T. T. Soe and F. R. Maxfield, J. Cell Biol., 1999, 144, 1271–1284 CrossRef CAS.
- H. T. McMahon and J. L. Gallop, Nature, 2005, 438, 590–596 CrossRef CAS PubMed.
- J. Zimmerberg and M. M. Kozlov, Nat. Rev. Mol. Cell Biol., 2006, 7, 9–19 CrossRef CAS PubMed.
- S. Vanni, L. Vamparys, R. Gautier, G. Drin, C. Etchebest, P. F. Fuchs and B. Antonny, Biophys. J., 2013, 104, 575–584 CrossRef CAS PubMed.
- N. S. Hatzakis, V. K. Bhatia, J. Larsen, K. L. Madsen, P. Y. Bolinger, A. H. Kunding, J. Castillo, U. Gether, P. Hedegard and D. Stamou, Nat. Chem. Biol., 2009, 5, 835–841 CrossRef CAS PubMed.
- B. J. Peter, H. M. Kent, I. G. Mills, Y. Vallis, P. J. Butler, P. R. Evans and H. T. McMahon, Science, 2004, 303, 495–499 CrossRef CAS PubMed.
- E. Casal, L. Federici, W. Zhang, J. Fernandez-Recio, E. M. Priego, R. N. Miguel, J. B. DuHadaway, G. C. Prendergast, B. F. Luisi and E. D. Laue, Biochemistry, 2006, 45, 12917–12928 CrossRef CAS PubMed.
- P. N. Dannhauser and E. J. Ungewickell, Nat. Cell Biol., 2012, 14, 634–639 CrossRef CAS PubMed.
- L. Vamparys, R. Gautier, S. Vanni, W. F. Bennett, D. P. Tieleman, B. Antonny, C. Etchebest and P. F. Fuchs, Biophys. J., 2013, 104, 585–593 CrossRef CAS PubMed.
- E. E. Kooijman, V. Chupin, N. L. Fuller, M. M. Kozlov, B. de Kruijff, K. N. Burger and P. R. Rand, Biochemistry, 2005, 44, 2097–2102 CrossRef CAS PubMed.
- M. M. Kamal, D. Mills, M. Grzybek and J. Howard, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 22245–22250 CrossRef CAS PubMed.
- W. B. Huttner and J. Zimmerberg, Curr. Opin. Cell Biol., 2001, 13, 478–484 CrossRef CAS.
- M. Heinrich, A. Tian, C. Esposito and T. Baumgart, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7208–7213 CrossRef CAS PubMed.
- H. Jiang, Phys. Rev. Lett., 2012, 109, 198101 CrossRef.
- F. Dumas, M. M. Sperotto, M. C. Lebrun, J. F. Tocanne and O. G. Mouritsen, Biophys. J., 1997, 73, 1940–1953 CrossRef CAS.
- A. Roux, D. Cuvelier, P. Nassoy, J. Prost, P. Bassereau and B. Goud, EMBO J., 2005, 24, 1537–1545 CrossRef CAS PubMed.
- M. Sundh, S. Svedhem and D. S. Sutherland, J. Phys. Chem. B, 2011, 115, 7838–7848 CrossRef CAS PubMed.
- S. F. Gilmore, H. Nanduri and A. N. Parikh, PLoS One, 2011, 6, e28517 CAS.
- A. H. Kunding, M. W. Mortensen, S. M. Christensen and D. Stamou, Biophys. J., 2008, 95, 1176–1188 CrossRef CAS PubMed.
- K. A. Sochacki, B. T. Larson, D. C. Sengupta, M. P. Daniels, G. Shtengel, H. F. Hess and J. W. Taraska, Nat. Commun., 2012, 3, 1154 CrossRef PubMed.
- J. W. Taraska, D. Perrais, M. Ohara-Imaizumi, S. Nagamatsu and W. Almers, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 2070–2075 CrossRef CAS PubMed.
- P. S. Cremer and S. G. Boxer, J. Phys. Chem. B, 1999, 103, 2554–2559 CrossRef CAS.
- L. K. Tamm, Biochemistry, 1988, 27, 1450–1457 CrossRef CAS.
- J. T. Groves, N. Ulman and S. G. Boxer, Science, 1997, 275, 651–653 CrossRef CAS.
- I. F. Sbalzarini and P. Koumoutsakos, J. Struct. Biol., 2005, 151, 182–195 CrossRef CAS PubMed.
- S. Barg, M. K. Knowles, X. Chen, M. Midorikawa and W. Almers, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20804–20809 CrossRef CAS PubMed.
- M. K. Knowles, S. Barg, L. Wan, M. Midorikawa, X. Chen and W. Almers, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20810–20815 CrossRef CAS PubMed.
- S. V. Costes, D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis and S. Lockett, Biophys. J., 2004, 86, 3993–4003 CrossRef CAS PubMed.
- P. Jonsson, M. P. Jonsson and F. Hook, Nano Lett., 2010, 10, 1900–1906 CrossRef PubMed.
- A. Edelstein, N. Amodaj, K. Hoover, R. Vale and N. Stuurman, Current Protocols in Molecular Biology, 2010, pp. 14.20.11–14.20.17 Search PubMed.
- D. Axelrod, D. E. Koppel, J. Schlessinger, E. Elson and W. W. Webb, Biophys. J., 1976, 16, 1055–1069 CrossRef CAS.
- M. Sundh, M. Manandhar, S. Svedhem and D. S. Sutherland, IEEE Trans. Nanobioscience, 2011, 10, 187–193 CrossRef PubMed.
- S. Mornet, O. Lambert, E. Duguet and A. Brisson, Nano Lett., 2005, 5, 281–285 CrossRef CAS PubMed.
- Y. Roiter, M. Ornatska, A. R. Rammohan, J. Balakrishnan, D. R. Heine and S. Minko, Nano Lett., 2008, 8, 941–944 CrossRef CAS PubMed.
- Y. Roiter, M. Ornatska, A. R. Rammohan, J. Balakrishnan, D. R. Heine and S. Minko, Langmuir, 2009, 25, 6287–6299 CrossRef CAS PubMed.
- W. Almers, Annu. Rev. Physiol., 1990, 52, 607–624 CrossRef CAS PubMed.
- J. H. Hurley and B. Wendland, Cell, 2002, 111, 143–146 CrossRef CAS.
- B. Sorre, A. Callan-Jones, J. B. Manneville, P. Nassoy, J. F. Joanny, J. Prost, B. Goud and P. Bassereau, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5622–5626 CrossRef CAS PubMed.
- T. S. Ursell, W. S. Klug and R. Phillips, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13301–13306 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplemental Methods and Figures. See DOI: 10.1039/c3sm52522h |
| This journal is © The Royal Society of Chemistry 2014 |