Open Access Article
Open Access ArticlePairing of cholesterol with oxidized phospholipid species in lipid bilayers†
Himanshu
Khandelia
*a,
Bastien
Loubet
a,
Agnieszka
Olżyńska
b,
Piotr
Jurkiewicz
b and
Martin
Hof
b
aMEMPHYS-Centre for Biomembrane Physics, Department of Physics and Chemistry, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark. E-mail: hkhandel@memphys.sdu.dk; Fax: +45 65504048; Tel: +45 65504048
bJ. Heyrovský Institute of Physical Chemistry of ASCR, v.v.i., Dolejškova 3, Prague 8, CZ-18223, Czech Republic. E-mail: martin.hof@jh-inst.cas.cz; Fax: +420-286 582 307; Tel: +420-266 053 264
First published on 19th November 2013
Abstract
We claim that (1) cholesterol protects bilayers from disruption caused by lipid oxidation by sequestering conical shaped oxidized lipid species such as 1-palmitoyl-2-azelaoyl-sn-glycero-3-phosphocholine (PZPC) away from phospholipid, because cholesterol and the oxidized lipid have complementary shapes and (2) mixtures of cholesterol and oxidized lipids can self-assemble into bilayers much like lysolipid–cholesterol mixtures. The evidence for bilayer protection comes from molecular dynamics (MD) simulations and dynamic light scattering (DLS) measurements. Unimodal size distributions of extruded vesicles (LUVETs) made up of a mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and PZPC containing high amounts of PZPC are only obtained when cholesterol is present in high concentrations. In simulations, bilayers containing high amounts of PZPC become porous, unless cholesterol is also present. The protective effect of cholesterol on oxidized lipids has been observed previously using electron paramagnetic resonance (EPR) and electron microscopy imaging of vesicles. The evidence for the pairing of cholesterol and PZPC comes mainly from correlated 2-D density and thickness plots from simulations, which show that these two molecules co-localize in bilayers. Further evidence that the two molecules can cohabitate comes from self-assembly simulations, where we show that cholesterol-oxidized lipid mixtures can form lamellar phases at specific concentrations, reminiscent of lysolipid–cholesterol mixtures. The additivity of the packing parameters of cholesterol and PZPC explains their cohabitation in a planar bilayer. Oxidized lipids are ubiquitously present in significant amounts in high- and low-density lipoprotein (HDL and LDL) particles, diseased tissues, and in model phospholipid mixtures containing polyunsaturated lipids. Therefore, our hypothesis has important consequences for cellular cholesterol trafficking; diseases related to oxidized lipids, and to biophysical studies of phase behaviour of cholesterol-containing phospholipid mixtures.
Introduction
The motivation of this report comes first, from an X-ray diffraction experiment which detected domain formation in peroxidized polyunsaturated bilayers containing cholesterol,1 and second, from the hypothesis that an inverted conical molecule (e.g. cholesterol) and a conical molecule (e.g. lysolipid) should seek each other out and pair-up in order to pack together into planar membranes composed of otherwise predominantly cylindrical phospholipids. While addressing these issues, we stumble upon a novel way to explain the protective effect that cholesterol has on bilayers which are assaulted by disrupting agents such as lysolipids.2In 2005, Jacob and Mason1 detected the formation of cholesterol domains in bilayers composed of mixtures of polyunsaturated lipids and cholesterol. Specifically, an additional d-spacing close to 34 Å was detected, and was interpreted as thinner cholesterol containing regions in the lipid matrix, which also contained thicker, cholesterol-depleted regions enriched in phospholipids. Cholesterol, however, is inverted conical in shape, and like surfactants, cannot form bilayers or bilayer domains on its own, but may do so when coupled with a molecule of complementary shape, such as a lysolipid.3 Such a region in a membrane, enriched in cholesterol–lysolipid pairs, will be of a different thickness than the surrounding phospholipid bilayer, and this might explain some of the observations of Jacob and Mason, who observed that the smaller d-spacing was only observed in the presence of cholesterol. The natural question that ensues is: where does the lysolipid moiety come from in such an experiment? Peroxidized bilayers do not produce lysolipids, but do produce a large number of oxidized lipid products (OXPLs), the sheer number of which makes it impossible to characterize the composition of such a bilayer. PZPC is a typical end-product, OXPL, which is well characterized, found in high concentrations in living and diseased cells,4,5 and is implicated in several disease conditions (Fig. 1). PZPC consists of a palmitoyl tail, and a shortened, oxidized acyl tail with a pendant carboxyl group, which reorients to find the lipid–water interface when PZPC is mixed with POPC lipids in a model lipid bilayer6,7 (see Fig. 1). In this form, PZPC can be considered to have a very large head group, comprised of the phosphatidylcholine moiety, plus the partially reoriented azealoyl tail. Thus, PZPC acquires a lysolipid like conical shape in a bilayer (Fig. 1). A ternary mixture of phospholipids, cholesterol and PZPC is thus a simple and effective model to test the above hypothesis.
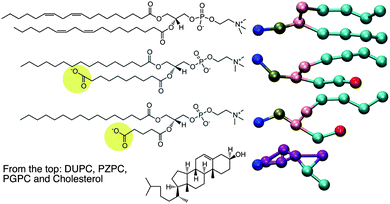 | ||
| Fig. 1 Left: the chemical structures of the molecules for which MD is carried out. On the right are the coarse-grained representations of the molecules in the MARTINI force field. | ||
Although we arrived at the above hypothesis independently during our investigations, it is by no means new. LysoPC–cholesterol mixtures form bilayers between 45 and 80% cholesterol (see ref. 3 and references therein). Kumar3 noted that although both cholesterol and lysoPCs have antagonistic effects on bilayers at high concentrations, they tend to cancel each other out when both are present in similar proportions. We investigate whether this pairing can also lead to formation of clusters or domains in bilayers. Cholesterol-pairing with lipids containing large head groups is also hypothesized in the so-called “Umbrella Model”.8
We have implemented coarse-grained Molecular Dynamics (CGMD) simulations of mixtures of DUPC (1,2-dilinoleoyl-sn-glycero-3-phosphocholine), PZPC and cholesterol to investigate possible lateral reorganization of the lipids resulting in formation of regions rich in both cholesterol and PZPC, and other DUPC-rich regions from which cholesterol and PZPC are excluded. We find that cholesterol and PZPC indeed are co-localized, but this pairing does not lead to explicit domain separation in the simulations. Our finding also explains the mechanism by which cholesterol protects bilayers from peroxidation.9 The protective effect of cholesterol has often been ascribed to its ordering effect on bilayers.10–12 We suggest that cholesterol protects bilayers from OXPLs by sequestering them into regions depleted in phospholipids, and are confident that a similar mechanism will also account for cholesterol's protective effects against lysolipids and perhaps other similarly shaped inverted cones.
The simulations results are complemented by Dynamic Light Scattering (DLS) measurements, which are performed to analyse the morphology of the model lipid membranes, composed of neutral DOPC lipids, oxidized PZPC lipids and cholesterol in various compositions. By measuring the size distribution of extruded liposomes (LUVETs) one can determine whether the size of vesicles is as expected after using this method of preparation (uniform and with the mean size similar to the pores of the membranes used for the extrusion), or if any alterations of vesicle morphology are present. Various types of alterations can be detected. It has been shown that an increased size of LUVETs can be caused by aggregation, which, e.g., might be the effect of the instability of the lamellar phase of lipids.13 In the case of LUVETs composed of more than one lipid species, decomposition of the vesicles can occur. This can be often detected using DLS, since the size distribution becomes multimodal in such a case.14
Our work has implications for the biomedical and biochemical sciences. OXPLs are known to affect membrane-associated phenomena, and are involved in a variety of disease conditions.5,15–18 Oxidized lipids have gained a lot from molecular simulations,19–23 because lipid oxidation produces innumerous transient intermediate compounds, which can be tedious to characterize in the lab biochemically and biophysically. Domain separation in models and real membranes is already recognized as a key research area in membrane biophysics, and there have been some reports on lateral reorganization in peroxidized cholesterol-free model membranes.24–27 But the problem becomes much richer when cholesterol is also present.
Results and discussion
Cholesterol protects bilayers from PZPC
Visual inspection is adequate to determine if each simulated bilayer is intact (able to separate two water compartments) after a few microseconds. Table 1 lists the “final results” from each simulation in the last column.| Ternary bilayer systems (cholesterol + PZPC + DUPC) | ||||
|---|---|---|---|---|
| Name | % CHOL | % oxidation | Onset of bilayer pores | Total simulation timea (μs) |
| a The time shown is actual simulated time multiplied by a factor of 4, to account for the faster sampling and thermal diffusion rates in coarse-grained simulations. b YY is the percentage of phospholipids that is oxidized. | ||||
| 0-YYb | 0 | 0 to 70% at 10% intervals | At 30% oxidation | 105 |
| 30-YY | 30 | 0 to 70% at 10% intervals | At 40% oxidation | 82 |
| 40-YY | 40 | 0 to 70% at 10% intervals | At 60% oxidation | 64 |
| Total ∼ 250 μs | ||||
| Binary self-assembly systems (cholesterol + PGPC) | |||
|---|---|---|---|
CHOL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGPC PGPC |
Number of simulations | % simulations in which bilayer forms | Total simulation time (μs) |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7 |
3 | 0 | 48 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
5 | 55.6 | 64 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
4 | 0 | 52 |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
9 | 0 | 184 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
3 | 0 | 48 |
| Total ∼ 400 μs | |||
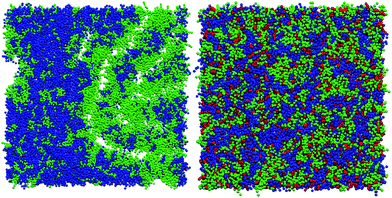
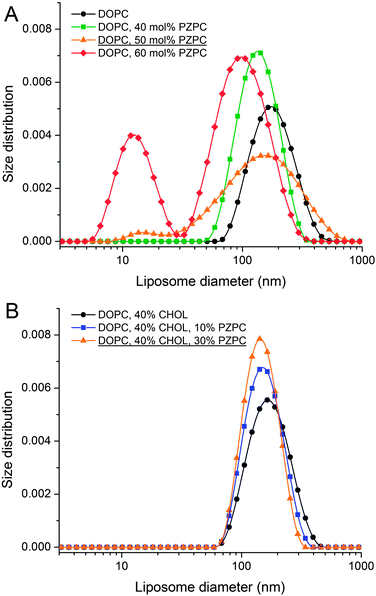
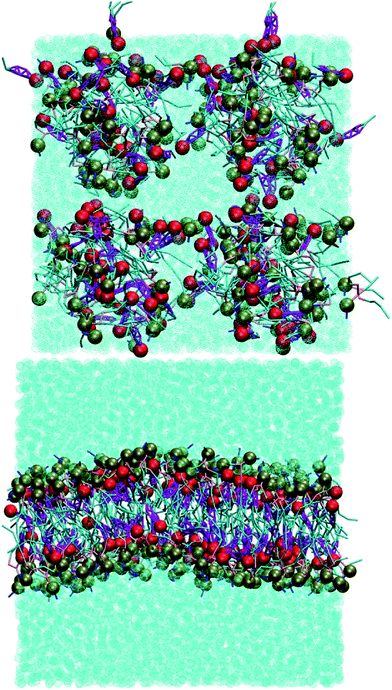
Without cholesterol, the bilayer first becomes porous at 30% oxidation. With 30% cholesterol, the bilayer first becomes porous at 40% oxidation. With 40% cholesterol, however, the bilayer is not porous even at 50% oxidation. Thus, increasing amounts of cholesterol protect the bilayer against increasing amounts of PZPC. A comparison of the final snapshots from the 0–50 (equimolar mixture of DUPC and PZPC) and the 40–50 (equimolar mixture of DUPC and PZPC, and 40 mol% cholesterol) is shown in Fig. 2. With the protonated version of PZPC, the bilayer first becomes porous at 40% oxidation. The protective role of cholesterol is apparent. The data are remarkably consistent with the DLS results.
In DLS, without cholesterol, a unimodal size distribution for each sample is observed up to 40% of PZPC (Fig. 3A). Note that transient or stable pores, or lateral homogeneity in the bilayer will not be detected by DLS if the vesicle size is unaffected. Beyond 50% PZPC, the distribution becomes bimodal (Fig. 3A), i.e. a second peak at ∼10 nm is present. We attribute this result to the decomposition of bilayers and formation of small PZPC-rich micelles. This finding supports the results of MD simulations. Samples containing 40 mol% cholesterol are characterized by unimodal size distribution up to 40 mol% of PZPC (Fig. 3B). The presence of 40 mol% cholesterol changes the size distribution of an equimolar DOPC–PZPC from bimodal to unimodal (compare Fig. 3A and B – equimolar samples are underlined). Apparently, cholesterol stabilizes the PZPC-containing bilayer.
Co-localization of PZPC and cholesterol
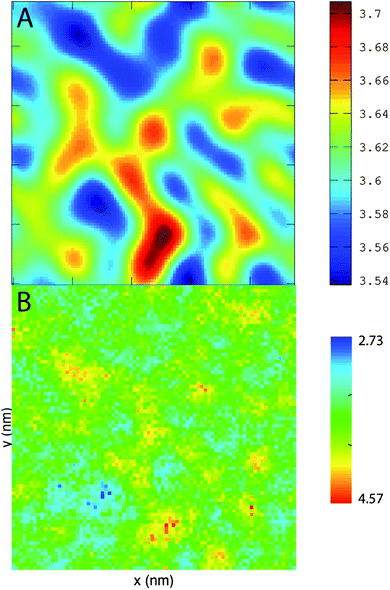
PZPC and cholesterol co-localize in the bilayer due to their complementary shapes. In Fig. 4, we show 2-D ensemble averaged density plots of individual molecules in the 40–50 simulation. DUPC is excluded from regions that are enriched in both cholesterol and PZPC. PZPC is clearly concentrated in regions where cholesterol is also present in high concentrations. The PZPC–cholesterol rich clusters are 1 to 4 nm in size, but do not acquire a well-defined morphology. Note that cholesterol is not completely excluded from the regions containing DUPC, but is only enriched locally where PZPC is present. Unlike in previous MARTINI-based coarse-grained simulations of archetypal phase-separating systems,28–33 we do not observe complete phase separation, but only small transient regions of the bilayer that are enriched in both cholesterol and PZPC. Surprisingly, the cholesterol–PZPC rich regions are thicker than the DUPC-rich regions, as evident in the 2D thickness profiles shown in Fig. 5. For example: the thinnest part of the bilayer at (x ∼ 13, y ∼ 3) is rich in DUPC. The minor variation in the thickness (from 3.54 to 3.7 nm) suggests that the domains are transient, and complete phase separation has not occurred.Further evidence of the formation of these “nano-domains” and the increased order in the bilayer comes from measuring the lateral 2-D diffusion coefficient (D) of individual molecules, which decreases nearly monotonically for all components in the mixtures (Fig. 6). Our numbers are of the same order of magnitude as the measured D of labelled OXPLs in a supported DOPC bilayer,34 and typical of diffusion coefficients in fluid bilayers.35 The decreasing D cannot be ascribed to an ordering of the bilayer induced by cholesterol for the highly oxidized systems, because the order parameters for the lipid tails are in fact lower in the 40–40 and 40–50 systems than in the non-oxidized 40–0 system (data not shown).
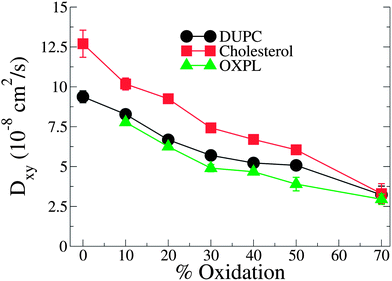 | ||
| Fig. 6 D lateral diffusion constants for cholesterol, PZPC and DUPC as a function of percentage of oxidation for systems containing 40% cholesterol. | ||
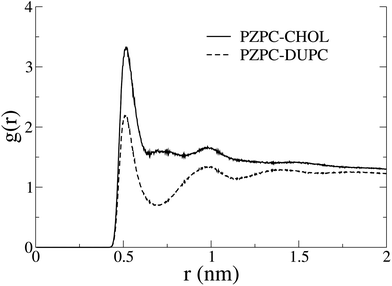
In Fig. 7 we show the radial distribution functions of PZPC with DUPC and cholesterol. Although the first and second peaks are identically located, interactions with cholesterol are stronger, and there is no minimum at 7 Å with cholesterol, suggesting that PZPC prefers to colocalize with cholesterol, than with DUPC.
If PZPC and cholesterol can co-localize in membranes, it is possible that these two molecules can form planar bilayers much like lysoPC–cholesterol mixtures.3 To investigate this possibility, we run self-assembly simulations of mixtures of cholesterol and a PZPC analogue: 1-palmitoyl-2-glutaroyl-sn-glycero-3-phosphocholine (PGPC), at various cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGPC ratios. The PGPC oxidized tail is shorter than PZPC by four methylene groups, corresponding to one MARTINI coarse-grained bead. We select PGPC to run the self-assembly simulations, in order to compare with EPR and electron microscopy experiments carried out on PGPC–cholesterol mixtures. We also run simulations with a protonated version of PGPC, which we call PGHC. PGPC is present in high concentrations in oxidatively modified low-density lipoprotein.36 We find that in 5 out of 9 simulation copies, cholesterol–PGHC mixtures form bilayers at a cholesterol
PGPC ratios. The PGPC oxidized tail is shorter than PZPC by four methylene groups, corresponding to one MARTINI coarse-grained bead. We select PGPC to run the self-assembly simulations, in order to compare with EPR and electron microscopy experiments carried out on PGPC–cholesterol mixtures. We also run simulations with a protonated version of PGPC, which we call PGHC. PGPC is present in high concentrations in oxidatively modified low-density lipoprotein.36 We find that in 5 out of 9 simulation copies, cholesterol–PGHC mixtures form bilayers at a cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGHC ratio of 2
PGHC ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. Bilayers are only formed with the protonated version of PGPC, and only at this specific ratio. Snapshots are shown in Fig. 8. Systems at all other compositions self-assemble into other microstructures such as micelles or bicelles. At CHOL
3. Bilayers are only formed with the protonated version of PGPC, and only at this specific ratio. Snapshots are shown in Fig. 8. Systems at all other compositions self-assemble into other microstructures such as micelles or bicelles. At CHOL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGHC 7
PGHC 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and 3
3 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, micelles are predominant, owing to the surfactant like shape of cholesterol. When the ratio is 1
2, micelles are predominant, owing to the surfactant like shape of cholesterol. When the ratio is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, bicelles are mostly formed and the bilayer is finally formed when the ratio becomes 2
1, bicelles are mostly formed and the bilayer is finally formed when the ratio becomes 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. The structures formed at 3
3. The structures formed at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 are aggregates of variable, undefined structures. Example simulation snapshots from each of these systems are shown in the ESI, Fig. S1.† The self-assembly simulation result is in wonderful quantitative agreement with Sepharose 4B gel-chromatography and cryo-electron microscopy,9 where lamellar structures were only observed at exactly a cholesterol
7 are aggregates of variable, undefined structures. Example simulation snapshots from each of these systems are shown in the ESI, Fig. S1.† The self-assembly simulation result is in wonderful quantitative agreement with Sepharose 4B gel-chromatography and cryo-electron microscopy,9 where lamellar structures were only observed at exactly a cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGHC ratio of 2
PGHC ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. The self-assembly of PGHC–cholesterol mixtures is strong evidence that oxidized phospholipids like PGPC and PZPC behave much like lysoPC. Density profiles from one of the self-assembled simulations are shown in the ESI, Fig. S2.† The carboxylate groups of the oxidized phospholipid and the cholesterol headgroup bead are at identical positions at the bilayer interface, suggesting a possible interaction between these chemical moieties, which however, cannot be further resolved in our CG simulations. The phosphate group of PGPC sits closer to the aqueous phase than the cholesterol headgroup. The non-zero density at the bilayer center for the COOH group shows that not all the COOH groups reverse their orientation in the bilayer. The non-zero density of the cholesterol headgroup at the bilayer center is a result of cholesterol flip-flop.
3. The self-assembly of PGHC–cholesterol mixtures is strong evidence that oxidized phospholipids like PGPC and PZPC behave much like lysoPC. Density profiles from one of the self-assembled simulations are shown in the ESI, Fig. S2.† The carboxylate groups of the oxidized phospholipid and the cholesterol headgroup bead are at identical positions at the bilayer interface, suggesting a possible interaction between these chemical moieties, which however, cannot be further resolved in our CG simulations. The phosphate group of PGPC sits closer to the aqueous phase than the cholesterol headgroup. The non-zero density at the bilayer center for the COOH group shows that not all the COOH groups reverse their orientation in the bilayer. The non-zero density of the cholesterol headgroup at the bilayer center is a result of cholesterol flip-flop.
Discussion
The contention of this report is that (1) cholesterol protects bilayers from disruption caused by lipid oxidation and (2) inverted-conical shaped cholesterol protects the bilayer by sequestering conical shaped oxidized lipid species away from the phospholipid, because cholesterol and the oxidized lipid have complementary shapes. We do not decline that the protective effect of cholesterol is also exerted by its ordering of the bilayer, as has been seen with other types of membrane-disrupting agents such as drugs and antimicrobial peptides,10–12 but argue that this may not be the dominant effect in some cases, such as disruption caused by molecules complementary in shape to cholesterol. For example, the order parameters of the lipid tails are lower in the oxidized bilayers (40–40 and 40–50) than in the non-oxidized system, and yet the bilayer is protected. To establish the above assertions, we have implemented about 700 μs of coarse-grained simulations and carried out simple but informative DLS measurements.The evidence for bilayer protection comes both from DLS and MD. Unimodal size distributions of extruded vesicles (LUVETs) made of DOPC–PZPC mixtures containing high amounts of PZPC are only obtained when cholesterol is present in high concentrations. Bilayers containing high amounts of PZPC become porous, unless cholesterol is also present. The most striking effect of cholesterol occurs at 50% oxidation of the bilayer. 40% cholesterol keeps these membranes intact, as indicated by both the DLS measurements, and the integrity of the bilayer in the MD simulations. The protective effect of cholesterol from oxidized lipids has previously been documented using EPR measurements coupled to images obtained from cyro-electron microscopy.9
The evidence for the pairing of cholesterol and PZPC comes from the 2-D density plots from simulations, which show that these two molecules co-localize in bilayers into regions that are thicker than those of the surrounding phospholipid-rich matrix. Furthermore, the lateral diffusion constant of all species in the bilayer mixtures decreases monotonically with increasing extents of oxidation. Further evidence that the two molecules can cohabitate comes from self-assembly simulations, where we show that cholesterol-oxidized lipid mixtures can form lamellar phases at specific concentrations, just like mixtures of cholesterol and lysolipids.3 The self-assembly of a cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGHC (2
PGHC (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) mixture into a bilayer is in very good agreement with cryo-electron microscopy images of vesicle formation by a mixture of identical composition.
3) mixture into a bilayer is in very good agreement with cryo-electron microscopy images of vesicle formation by a mixture of identical composition.
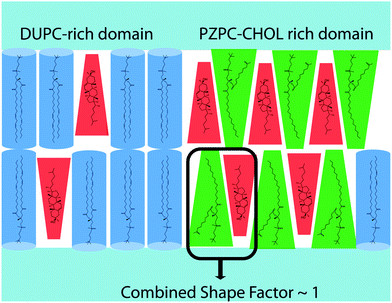
The pairing of molecules of conical and inverted conical shapes in bilayers can be understood if terms of the additivity of the packing parameter,3 defined as S = V/al, where V is the volume of the hydrocarbon volume, l is the length of the hydrocarbon tail, and a the area of the head group.37,38 Lysolipids and PZPC have a large head group (large V), and S > 1, and such molecules form micelles. Cholesterol has a much smaller head group, and S is below 1/2. Thus, cholesterol will adopt a hexagonal phase, like phosphatidylethanol lipids. However, when both cholesterol and PZPC are present, they can co-localize, and the additive value of S can be between 0.7 and 1, corresponding to the bilayer-forming regime of S.3 The additivity of the packing parameter is sufficient to explain the colocalization of PZPC and cholesterol in our simulations. Our hypothesis on the lateral organization of the ternary lipid mixture is illustrated in Fig. 9.
The reduced lateral diffusion coefficient (D) of labelled oxidized lipids in bilayers in the presence of cholesterol was measured in supported lipid bilayers.34 Additional MD simulations34 ascribed the decreasing D to the deeper location of the oxidized lipid headgroup at higher cholesterol concentrations. Our observations are similar with regard to the decreasing D, but we interpret the results differently as described above.
A slightly puzzling issue in the story is that we cannot very clearly detect different phases by means of either radial distribution functions in terms of an outright preference for cholesterol towards one region or another, or dynamically changing the length of the domain interface.28,29,31–33,39–41 We ascribe this anomaly to incomplete phase separation. The large-scale heterogeneity in the system masks smaller clusters of PZPC and cholesterol. The radial distribution functions in Fig. 7 do not emulate the compelling nature of the evidence from the 2D density plots and thickness profiles in Fig. 4 and 5. Complete phase separation is possibly driven by other phenomena that would come to the force only if additional degrees of freedom are present in our mixtures, for example: in the form of new molecules, as is probably the case with Jacob and Mason's experiments,1 where there are many different phospholipid oxidation products, and possibly cholesterol oxidation products may also be present. We do not think simulation timescales are a problem, because for most canonical phase separating mixtures, phase separation does occur on smaller time scales in MARTINI-based coarse-grained simulations.14,28,29,31–33,40,42
Some unresolved questions that still persist are: (1) why does the anionic form of PGPC not form bilayers with cholesterol at all? The uncharged form of PZPC is also less perturbing to the bilayer compared to the anionic form. Electrostatic repulsions between adjoining OXPL species are probably responsible for this effect. (2) What might be the possible role of oxidized cholesterol species in the behavior of these mixtures? (3) Why are PZPC-rich regions slightly thicker than the DUPC-rich regions in the ternary simulations? One possibility is that the entire headgroup of PZPC is pulled closer to the aqueous phase because of the chain reversal of the oxidized tail. We are currently addressing some of these questions in additional investigations.
Conclusions
We present a novel hypothesis of the preferential cohabitation of cholesterol and OXPLs in lipid bilayers, and also propose a new mechanism by which cholesterol may protect bilayers from disrupting agents. Our results are relevant for the health sciences and the membrane biophysics community involved in studying membrane domains in general. PZPC, and similarly shaped PGPC and other OXPLs are present in high concentrations in lipoprotein particles that are also rich in cholesterol.36 These OXPLs are also present in apoptotic tissues and are involved in a large variety of ailments, which are reviewed in ref. 43. Mammalian cell membranes contain between 20 and 50% cholesterol. Therefore the analysis of PZPC–cholesterol interactions is important. Our hypothesis suggests that cholesterol can not only protect mammalian membranes from PZPC, but can also sequester PZPC into cholesterol–PZPC rich regions in the membrane. Such clusters are also likely to form in LDL particles, and will likely affect the overall trafficking of cholesterol in the body. The phase diagrams of cholesterol-containing ternary phase-separating phospholipid mixtures may also need readjustment if there is clear evidence of the presence of oxidized phospholipid species such as PZPC in the mixture.Experimental
Simulations
All simulations are carried out in GROMACS v. 4.5.x44–47 using the MARTINI coarse-grained force field for lipids,48 because we anticipate packing to be the main driver of lateral reorganization, and therefore do not attempt to describe specific interactions atomistically. Of course, MARTINI allows simulations of longer time and length scales. A list of the implemented simulations is provided in Table 1. Simulations of bilayers containing various proportions of phospholipid, PZPC and cholesterol are implemented. The model phospholipid used is 1,2-dilinoleoyl-sn-glycero-3-phosphocholine (DUPC), the same as in Jacob and Mason's experiments, but our main conclusions will apply to any typical model phospholipid such as DOPC or POPC. The percentage of oxidation ranged from 0 to 70% in 10% intervals, while cholesterol was simulated at 0, 30 and 40% molar percentages. We annotate the simulations as XX–YY, where XX is the total percentage of cholesterol present, and YY is the percentage of phospholipids that are oxidized. For example, for a 100-lipid system, 40–30 refers to a system containing 40 molecules of cholesterol, 21 molecules of PZPC and 39 molecules of DUPC. The total number of resulting simulations is 24. Simulations with 0, 30, and 40% cholesterol and 20, 30 and 40% PZPC are repeated with a protonated version of PZPC, and the results are identical in most respects. We therefore only report results from the deprotonated version for brevity.We start with a 29 nm × 29 nm DUPC bilayer patch containing 2250 lipids, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 MARTINI water molecules corresponding to 26.4 water molecules per lipid. For systems containing PZPC, we also add an equal number of Na+ ions to keep the simulation box electrostatically neutral. Simulations are run for 1 to 5 μs MARTINI time, corresponding to 4–20 μs real simulation time for each simulation. A leapfrog algorithm with a time step of 20 fs was used. Default parameters for non-bonded interactions recommended in the MARTINI force field are used. Simulations are run in the NPT ensemble at 310 K and 1 atm, with a Berendsen thermo- and barostat.49 The coupling time constants are 5 ps and 3 ps for P and T respectively. The aggregate simulation time exceeds 250 μs.
850 MARTINI water molecules corresponding to 26.4 water molecules per lipid. For systems containing PZPC, we also add an equal number of Na+ ions to keep the simulation box electrostatically neutral. Simulations are run for 1 to 5 μs MARTINI time, corresponding to 4–20 μs real simulation time for each simulation. A leapfrog algorithm with a time step of 20 fs was used. Default parameters for non-bonded interactions recommended in the MARTINI force field are used. Simulations are run in the NPT ensemble at 310 K and 1 atm, with a Berendsen thermo- and barostat.49 The coupling time constants are 5 ps and 3 ps for P and T respectively. The aggregate simulation time exceeds 250 μs.
We also implement self-assembly simulations of mixtures of cholesterol and a PZPC analogue: 1-palmitoyl-2-glutaroyl-sn-glycero-3-phosphocholine (PGPC), at various cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PGPC ratios. Although the exact ratios at which PZPC–cholesterol and PGHC (or PGPC)–cholesterol mixtures form bilayers may differ a bit, the fundamental driving force of shape matching is identical. The mixtures contain 300 lipids and 4800 MARTINI water beads. The proportion of cholesterol is varied from 30 to 70% in 10% intervals. Simulation parameters are similar to the bilayer simulations. The aggregate simulation time for the self-assembly simulations exceeds 400 μs.
PGPC ratios. Although the exact ratios at which PZPC–cholesterol and PGHC (or PGPC)–cholesterol mixtures form bilayers may differ a bit, the fundamental driving force of shape matching is identical. The mixtures contain 300 lipids and 4800 MARTINI water beads. The proportion of cholesterol is varied from 30 to 70% in 10% intervals. Simulation parameters are similar to the bilayer simulations. The aggregate simulation time for the self-assembly simulations exceeds 400 μs.
Dynamic light scattering
Preparation of lipid bilayers
The required volumes of chloroform solutions of DOPC, PZPC and cholesterol were mixed in a glass tube. Solvents were evaporated under a stream of nitrogen. The lipid film was kept in vacuum for at least 2 hours. The dry lipid film was then suspended in 10 mM HEPES buffer (pH = 7.4, 150 mM NaCl) and vortexed for 5 minutes. The final total concentration of lipids with (or without) cholesterol in the sample was 0.2 mM. Extruded large unilamellar vesicles (LUVETs) were formed by extrusion through a polycarbonate membrane (Avestin, Ottawa, Canada) of a nominal pore diameter of 100 nm. All prepared samples were transferred to 1 cm plastic cuvettes and equilibrated at 25 °C for 3 minutes before each measurement.DLS measurements
Dynamic light scattering (DLS) was used to measure vesicle size distribution. The light scattering setup (Zetasizer Nano ZS, Worcestershire, UK) consisted of a He–Ne laser 532 nm and an avalanche photodiode detector (APD). The scattering intensity was collected at the angle of 173°. Using cumulant analysis, the mean vesicle size (Z-average) for every monodisperse sample was obtained. The systems analysed are a cholesterol-free DOPC bilayer containing increasing amount of PZPC up to 70 mol% and mixtures of 40 mol% cholesterol and DOPC with increasing content of PZPC up to 66%.Acknowledgements
The computations were done at the SDU node of the Danish Centre for Scientific Computing (DCSC). HK is funded by a Lundbeck Junior Group Leader Investigator Fellowship. AO, PJ, and MH acknowledge the Grant Agency of the Czech Republic (P106/12/G016). MH acknowledges the Praemium Academie Award. We thank Albin Hermetter and Francesco Megli for useful discussions.Notes and references
- R. F. Jacob and R. P. Mason, J. Biol. Chem., 2005, 280, 39380–39387 CrossRef PubMed.
- K. W. Liu and S. L. Biswal, Anal. Chem., 2011, 83, 4794–4801 CrossRef PubMed.
- V. V. Kumar, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 444–448 CrossRef.
- S. S. Davies, A. V. Pontsler, G. K. Marathe, K. A. Harrison, R. C. Murphy, J. C. Hinshaw, G. D. Prestwich, A. S. Hilaire, S. M. Prescott, G. A. Zimmerman and T. M. McIntyre, J. Biol. Chem., 2001, 276, 16015–16023 CrossRef PubMed.
- G. O. Fruhwirth, A. Loidl and A. Hermetter, Biochim. Biophys. Acta, Mol. Basis Dis., 2007, 1772, 718–736 CrossRef PubMed.
- H. Khandelia and O. G. Mouritsen, Biophys. J., 2009, 96, 2734–2743 CrossRef PubMed.
- M. E. Greenberg, X. M. Li, B. G. Gugiu, X. D. Gu, J. Qin, R. G. Salomon and S. L. Hazen, J. Biol. Chem., 2008, 283, 2385–2396 CrossRef PubMed.
- J. Huang and G. W. Feigenson, Biophys. J., 1999, 76, 2142–2157 CrossRef.
- F. M. Megli, E. Conte and T. Ishikawa, Biochim. Biophys. Acta, 2011, 1808, 2267–2274 CrossRef PubMed.
- J. Henriksen, A. C. Rowat, E. Brief, Y. W. Hsueh, J. L. Thewalt, M. J. Zuckermann and J. H. Ipsen, Biophys. J., 2006, 90, 1639–1649 CrossRef PubMed.
- A. J. McHenry, M. F. M. Sciacca, J. R. Brender and A. Ramamoorthy, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 3019–3024 CrossRef PubMed.
- C. L. Wennberg, D. van der Spoel and J. S. Hub, J. Am. Chem. Soc., 2012, 134, 5351–5361 CrossRef PubMed.
- A. Di Biasio, F. Bordi and C. Cametti, Prog. Colloid Polym. Sci., 2004, 123, 78–82 Search PubMed.
- R. F. Correia, M. I. Viseu, T. J. V. Prazeres and J. M. G. Martinho, J. Colloid Interface Sci., 2012, 379, 56–63 CrossRef PubMed.
- U. Stemmer, C. Ramprecht, E. Zenzmaier, B. Stojcic, G. Rechberger, M. Kollroser and A. Hermetter, Biochim. Biophys. Acta, 2012, 1821, 706–718 CrossRef PubMed.
- A. Hermetter, P. Kinnunen and C. Spickett, Biochim. Biophys. Acta, 2012, 1818, 2373 CrossRef PubMed.
- U. Stemmer, Z. A. Dunai, D. Koller, G. Purstinger, E. Zenzmaier, H. P. Deigner, E. Aflaki, D. Kratky and A. Hermetter, Lipids Health Dis., 2012, 11, 110 CrossRef PubMed.
- T. M. McIntyre, Biochim. Biophys. Acta, 2012, 1818, 2456–2464 CrossRef PubMed.
- P. Jurkiewicz, A. Olzynska, L. Cwiklik, E. Conte, P. Jungwirth, F. M. Megli and M. Hof, Biochim. Biophys. Acta, 2012, 1818, 2388–2402 CrossRef PubMed.
- L. Beranova, L. Cwiklik, P. Jurkiewicz, M. Hof and P. Jungwirth, Langmuir, 2010, 26, 6140–6144 CrossRef PubMed.
- M. Vazdar, P. Jurkiewicz, M. Hof, P. Jungwirth and L. Cwiklik, J. Phys. Chem. B, 2012, 116, 6411–6415 CrossRef PubMed.
- J. Wong-Ekkabut, Z. Xu, W. Triampo, I. M. Tang, D. P. Tieleman and L. Monticelli, Biophys. J., 2007, 93, 4225–4236 CrossRef CAS PubMed.
- A. Hermetter, W. Kopec and H. Khandelia, Biochim. Biophys. Acta, 2013, 1828, 1700–1706 CrossRef CAS PubMed.
- F. M. Megli, L. Russo and K. Sabatini, FEBS Lett., 2005, 579, 4577–4584 CrossRef CAS PubMed.
- R. P. Mason, M. F. Walter and P. E. Mason, Free Radical Biol. Med., 1997, 23, 419–425 CrossRef CAS.
- U. Muscatello, A. Alessandrini, G. Valdre, V. Vannini and U. Valdre, Biochem. Biophys. Res. Commun., 2000, 270, 448–452 CrossRef CAS PubMed.
- J. Zhao, J. Wu, H. L. Shao, F. Kong, N. Jain, G. Hunt and G. Feigenson, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 2777–2786 CrossRef CAS PubMed.
- S. Baoukina, E. Mendez-Villuendas, W. F. D. Bennett and D. P. Tieleman, Faraday Discuss., 2013, 161, 63–75 RSC.
- H. J. Risselada and S. J. Marrink, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17367–17372 CrossRef CAS PubMed.
- Q. Waheed, R. Tjornhammar and O. Edholm, Biophys. J., 2012, 103, 2125–2133 CrossRef CAS PubMed.
- C. Rosetti and C. Pastorino, J. Phys. Chem. B, 2012, 116, 3525–3537 CrossRef CAS PubMed.
- D. Hakobyan and A. Heuer, J. Phys. Chem. B, 2013, 117, 3841–3851 CrossRef CAS PubMed.
- R. S. Davis, P. B. S. Kumar, M. M. Sperotto and M. Laradji, J. Phys. Chem. B, 2013, 117, 4072–4080 CrossRef CAS PubMed.
- B. Plochberger, T. Stockner, S. Chiantia, M. Brameshuber, J. Weghuber, A. Hermetter, P. Schwille and G. J. Schutz, Langmuir, 2010, 26, 17322–17329 CrossRef CAS PubMed.
- G. Lindblom and G. Oradd, Prog. Nucl. Magn. Reson. Spectrosc., 1994, 26, 483–515 CrossRef CAS.
- G. O. Fruhwirth, A. Moumtzi, A. Loidl, E. Ingolic and A. Hermetter, Biochim. Biophys. Acta, 2006, 1761, 1060–1069 CrossRef CAS PubMed.
- J. N. Israelachvili and D. J. Mitchell, Biochim. Biophys. Acta, 1975, 389, 13–19 CrossRef CAS.
- J. N. Israelachvili, D. J. Mitchell and B. W. Ninham, J. Chem. Soc., Faraday Trans. 2, 1976, 72, 1525–1568 RSC.
- P. W. Tumaneng, S. A. Pandit, G. J. Zhao and H. L. Scott, J. Chem. Phys., 2010, 132, 065104 CrossRef PubMed.
- D. V. Pyrkova, N. K. Tarasova, N. A. Krylov, D. E. Nolde and R. G. Efremov, Biol. Membr., 2011, 28, 298–306 CAS.
- P. S. Niemela, S. Ollila, M. T. Hyvonen, M. Karttunen and I. Vattulainen, PLoS Comput. Biol., 2007, 3, e34 Search PubMed.
- J. de Joannis, P. S. Coppock, F. C. Yin, M. Mori, A. Zamorano and J. T. Kindt, J. Am. Chem. Soc., 2011, 133, 3625–3634 CrossRef CAS PubMed.
- P. K. J. Kinnunen, K. Kaarniranta and A. K. Mahalka, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 2446–2455 CrossRef CAS PubMed.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS.
- E. Lindahl, B. Hess and D. van der Spoel, J. Mol. Model., 2001, 7, 306–317 CAS.
- D. V. D. Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef PubMed.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. de Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Vangunsteren, A. Dinola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS PubMed.
- N. A. Porter, L. S. Lehman, B. A. Weber and K. J. Smith, J. Am. Chem. Soc., 1981, 103, 6447–6455 CrossRef CAS.
- T. M. McIntyre, G. A. Zimmerman and S. M. Prescott, J. Biol. Chem., 1999, 274, 25189–25192 CrossRef CAS PubMed.
- S. Parthasarathy, J. C. Khoo, E. Miller, J. Barnett, J. L. Witztum and D. Steinberg, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 3894–3898 CrossRef CAS.
- D. Marsh, Biophys. J., 1997, 72, 2834–2836 CrossRef CAS.
- J. N. Israelachvili, D. J. Mitchell and B. W. Ninham, Biochim. Biophys. Acta, 1977, 470, 185–201 CrossRef CAS.
- N. Kucerka, J. F. Nagle, J. N. Sachs, S. E. Feller, J. Pencer, A. Jackson and J. Katsaras, Biophys. J., 2008, 95, 2356–2367 CrossRef CAS PubMed.
- M. Stepniewski, A. Bunker, M. Pasenkiewicz-Gierula, M. Karttunen and T. Rog, J. Phys. Chem. B, 2010, 114, 11784–11792 CrossRef CAS PubMed.
- W. Rawicz, K. C. Olbrich, T. McIntosh, D. Needham and E. Evans, Biophys. J., 2000, 79, 328–339 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3sm52310a |
| This journal is © The Royal Society of Chemistry 2014 |