Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Time-resolved botulinum neurotoxin A activity monitored using peptide-functionalized Au nanoparticle energy transfer sensors†
Yi
Wang
ab,
Xiaohu
Liu
a,
Jinling
Zhang
ab,
Daniel
Aili
ac and
Bo
Liedberg
*ab
aCentre for Biomimetic Sensor Science, Nanyang Technological University, 50 Nanyang Drive, 637553 Singapore. E-mail: bliedberg@ntu.edu.sg; Tel: +65 6316 2957
bSchool of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, 639798 Singapore
cDivision of Molecular Physics, Department of Physics, Chemistry and Biology, Linköping University, 58183 Linköping, Sweden
First published on 26th February 2014
Abstract
We report herein on the employment of synthetic peptide-functionalized gold nanoparticles (AuNPs) with various diameters as radiative quenchers for the time-resolved monitoring of botulinum A light chain (BoLcA) activity. The results demonstrate that larger AuNPs provide higher energy transfer efficiencies between the dye and the AuNPs, but poorer BoLcA activities for the proteolysis of peptides because of steric constraints. The initial turnover number for the BoLcA proteolysis of peptides on 18 nm AuNPs was retarded by a factor of 80 as compared with 1.4 nm AuNPs. A similar phenomenon has been observed for trypsin, however, with less hindrance on large AuNPs. Thus, the use of smaller 1.4 nm AuNPs in conjunction with robust synthetic peptides provides an attractive format for the time-resolved monitoring of protease activity and for BoLcA sensing at a highly competitive limit of detection (1 pM).
Introduction
Botulinum neurotoxins (BoNTs), produced by Clostridium botulinum,1 are considered to be the most lethal substance known to humans. BoNTs are classified into seven immunologically distinct serotypes (A–G) each of which can cause flaccid muscle paralysis and subsequent death by blocking the release of neurotransmitters at neuromuscular junctions. Structurally, the neurotoxins are expressed as a single chain polypeptide which after post-translational proteolysis consists of two subunits: a 100 kDa heavy chain (HC) and a 50 kDa light chain (LC) linked via a disulfide bond. The HC is responsible for the binding and translocation of the toxin across the synaptic membrane through specific receptors; whereas the LC functions as an active zinc-endopeptidase that cleaves the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins leading to the inhibition of acetylcholine release and subsequent neuroparalysis.2 The neurotoxin can enter the body via the gastrointestinal tract or through mucous membranes of, for instance, the eyes or the respiratory tract. In humans, a lethal dose intravenously is estimated at 1–2 ng kg−1 body weight, orally at 1 μg kg−1 and 10–12 ng kg−1 by inhalation.3 The gold standard “mouse bioassay” is able to detect as little as 10 pg ml−1 of toxin,4 however, it requires several days of assay time, a large number of animals and can only be performed at specific laboratories. Therefore rapid, sensitive and easily accessible assays are required to meet biodefense diagnostic and therapeutic needs.A number of in vitro assays to detect BoNTs, including ELISA,5 immuno-PCR,6 surface plasmon resonance (SPR) immunoassay7 and electrochemical luminescence8 have been developed.9 However, these assays do not usually provide information on the enzymatic activity that is responsible for the toxicity of BoNTs. By rational design of a peptide substrate containing the BoNTs cleavage site derived from the SNARE protein, it is possible to monitor the activity of various serotypes of the BoNTs as each serotype cleaves at a different site on the SNARE protein. Assays to monitor the activity of BoNTs are typically based on measuring mass changes after the cleavage of the substrate via SPR;10 counting the number of amplifiers or labels such as phage-based amplifiers,11 or monitoring the reduction in the fluorescence resonance energy transfer (FRET) upon the cleavage of substrates.12 Among these methods FRET has been extensively applied for the monitoring of proteolytic activities of various proteases including trypsin, caspase 1, caspase 3, thrombin, chymotrypsin, collagenase, HIV-1 protease and so on.13 Several of the organic fluorophores commonly used in FRET bioassays suffer from pH sensitivity, photo-bleaching, chemical degradation and the overlapping of absorption spectra between the acceptor and donor. To address these problems, quantum dots (QDs) have been employed in FRET measurements because of their relative stability, tuneable emissions for the optimization of spectral overlap with a particular acceptor, and potential for minimization of the direct acceptor excitation.14 However, the overlap between the emissions of the donor and acceptor is still unavoidable, in addition to the potential toxicity of QDs.15 Accordingly, having an acceptor that is non-fluorescent, such as metallic nanoparticles, can help to solve these issues. Metal nanoparticles have found applications either as radiative quenchers or radiative enhancers, depending on the particle size, shape, composition, and the distance between the donor and the metal nanoparticle.16 For Au nanoparticles smaller than 40 nm in diameter, the absorption term dominates over the scattering term, and radiative quenching becomes the main cause for energy transfer from the dye to the metal which results in electron–hole pair formations and subsequent Ohmic losses.16a On the contrary, large nanoparticles are expected to enhance the fluorescence because scattering dominates over absorption. Au nanoparticles have been previously utilized as quenching beacons for the detection of DNA,17 RNA,18 proteases19 and other biomolecules.20
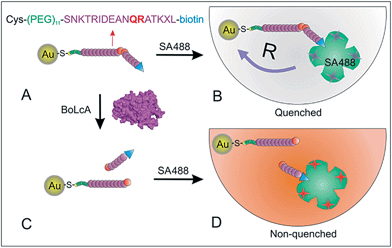
In this paper, Au nanoparticles with diameters of 1.4 nm, 6 nm and 18 nm were modified with peptides containing a BoLcA cleavage site (AuNP–pep) and employed as radiative quenchers for monitoring the BoLcA activity. The AuNP–pep construct was pre-treated (cleaved) with BoLcA prior to incubation with the streptavidin–Alexa488 conjugate (SA488) to enable the measurement of the fluorescence intensity (Fig. 1). The fluorescence energy transfer efficiency was first investigated for AuNPs of different sizes. Then, the time-resolved catalytic activity of BoLcA on the AuNP–pep constructs was monitored and compared with the activity of trypsin on the same AuNP–pep construct. Finally, we employed the assay method shown in Fig. 1 for BoLcA sensing.
Results and discussion
Peptide sequence
Previous research has reported that a short peptide derived from the C-terminal end (187–203) of the SNAP-25 protein (part of the SNARE complex) can be hydrolyzed by BoNTs at a similar rate as the full length SNAP-25, as long as the peptide has a minimum length of 14 to 16 amino acids and arginine is present at the cleavage site.21 The peptide sequence used in our study was based on the 17 amino acids at the C-terminus of the SNAP-25 protein (187–203). The glutamine–arginine (Q–R) located in the middle of the target peptide is the cleavage site for BoLcA proteolysis according to mass spectrometer results (Fig. S1†) and previous work.21,22 Furthermore, substitution of methionine (M) in position 202 with norleucine (X) increases the proteolytic rate of BoLcA.21,23 At the N-terminus of the peptide a cysteine was attached to enable its immobilization on the AuNPs via the –SH moiety.24 In addition, an 11 unit ethylene glycol oligomer was introduced as a spacer between the Cys group and the recognition sequence to minimize the steric hindrance caused by the AuNPs. At the C-teriminus a biotin molecule was added to enable the specific interaction with the streptavidin–dye conjugate. SA488 contains 3–6 dyes per protein (see ESI†), which is advantageous compared with a single dye-labelled peptide, as it provides a higher fluorescence response to the peptide cleavage-induced dye release.AuNP size-dependent energy transfer efficiency
The peptide was attached to AuNPs with diameters of 1.4, 6 and 18 nm (see experimental details in ESI and Fig. S2†) and their respective extinction spectra are shown in Fig. 2A. The 1.4 nm AuNPs displayed significantly lower extinction coefficients as compared with the 6 nm and 18 nm AuNPs because of their low electron density at the conduction band and large damping effect.25 The 1.4 nm AuNP modified with the peptide substrate (AuNP–pep) was then incubated with SA488 at different molar ratios. The fluorescence intensity at λ = 520 nm decreased from I0 = 5.45 × 105 cps μA−1 for 10 nM SA488 to I′ = 2.0 × 105 cps μA−1 upon increasing the molar ratio of AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 to 20
SA488 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2B), and the energy transfer efficiency E = 1 − I′/I0 became saturated at 62.5% for a AuNP–pep
1 (Fig. 2B), and the energy transfer efficiency E = 1 − I′/I0 became saturated at 62.5% for a AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 ratio ≥ 5
SA488 ratio ≥ 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 2C). The maximum fluorescence quenching efficiency was observed for a AuNP–pep
1 (Fig. 2C). The maximum fluorescence quenching efficiency was observed for a AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 ratio of ∼5
SA488 ratio of ∼5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 which is reasonable given that streptavidin has 4 affinity binding sites for biotins. Previous work suggests that small AuNPs with diameters of ∼1–3 nm as acceptors can be described by the nanometal surface energy transfer (NSET) theory.16a In contrast to FRET, NSET does not require a resonant electronic transition as it originates from the interaction of the electromagnetic field of the donor dipole with the free conduction electrons of the accepting metal. The NSET characteristic distance R0 is expressed as
1 which is reasonable given that streptavidin has 4 affinity binding sites for biotins. Previous work suggests that small AuNPs with diameters of ∼1–3 nm as acceptors can be described by the nanometal surface energy transfer (NSET) theory.16a In contrast to FRET, NSET does not require a resonant electronic transition as it originates from the interaction of the electromagnetic field of the donor dipole with the free conduction electrons of the accepting metal. The NSET characteristic distance R0 is expressed as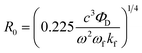 | (1) |
 | (2) |
As compared with 1.4 nm AuNPs, the larger AuNPs, with diameters of 6 nm and 18 nm, displayed increased fluorescence quenching efficiencies (Fig. 2C). The fluorescence quenching efficiencies for 6 nm and 18 nm AuNPs reached a maximum of around 90% and 98% for molar ratios of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, respectively. This is reasonable as the large AuNPs offer higher extinction coefficients and carry a larger number of peptides. The 1.4 nm AuNPs were modified with a single maleimide group for the attachment of one peptide per AuNP (see ESI†). The average number of peptides on each 18 nm and 6 nm AuNP was estimated to be 51 and 3, respectively. This estimation is based on a peptide/thiol-PEG molar ratio of 1
3, respectively. This is reasonable as the large AuNPs offer higher extinction coefficients and carry a larger number of peptides. The 1.4 nm AuNPs were modified with a single maleimide group for the attachment of one peptide per AuNP (see ESI†). The average number of peptides on each 18 nm and 6 nm AuNP was estimated to be 51 and 3, respectively. This estimation is based on a peptide/thiol-PEG molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (18 nm AuNPs) and 0.5
100 (18 nm AuNPs) and 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (6 nm AuNPs), respectively, and the assumption that each thiol occupies an area of 0.2 nm2. These observations, shown in Fig. 2, suggest that the fluorescence quenching efficiency is determined by both the size of the AuNPs, and the coverage of peptides on the AuNPs. The fluorescence quenching efficiency reached a maximum at a lower AuNP–pep
100 (6 nm AuNPs), respectively, and the assumption that each thiol occupies an area of 0.2 nm2. These observations, shown in Fig. 2, suggest that the fluorescence quenching efficiency is determined by both the size of the AuNPs, and the coverage of peptides on the AuNPs. The fluorescence quenching efficiency reached a maximum at a lower AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 ratio when the coverage of peptide on 6 nm AuNPs was increased (Fig. S3†). Furthermore, the energy transfer for large AuNPs with diameters of 6 nm and 18 nm was assumed to be dominated by FRET, from which the characteristic radius (R0) was estimated as R0 = 16.6 and 29.8 nm, respectively (see ESI†). Accordingly, based on the calculation that the distance between the dye and the AuNP surface is about R = 6.85 nm, we estimate a quenching efficiency of E ∼ 95% and 98% for the 6 and 18 nm AuNPs, respectively (eqn (2)). These values are consistent with the maximum fluorescence quenching efficiencies of E = 92% and 98% for the 6 and 18 nm AuNPs, respectively, as presented in Fig. 2C. Thus, our results confirm that the energy transfer on AuNPs with a diameter larger than 6 nm follows the FRET mechanism. One can of course argue that there is always a risk for aggregation between AuNPs induced by the multivalency of streptavidin (4 binding pockets per molecule). This might occur when there is more than one peptide per AuNP (as for the 6 and 18 nm particles). However, no colour change nor any plasmon band shift were observed after mixing SA488 with the 6 nm AuNPs at the molar ratio of AuNP–pep
SA488 ratio when the coverage of peptide on 6 nm AuNPs was increased (Fig. S3†). Furthermore, the energy transfer for large AuNPs with diameters of 6 nm and 18 nm was assumed to be dominated by FRET, from which the characteristic radius (R0) was estimated as R0 = 16.6 and 29.8 nm, respectively (see ESI†). Accordingly, based on the calculation that the distance between the dye and the AuNP surface is about R = 6.85 nm, we estimate a quenching efficiency of E ∼ 95% and 98% for the 6 and 18 nm AuNPs, respectively (eqn (2)). These values are consistent with the maximum fluorescence quenching efficiencies of E = 92% and 98% for the 6 and 18 nm AuNPs, respectively, as presented in Fig. 2C. Thus, our results confirm that the energy transfer on AuNPs with a diameter larger than 6 nm follows the FRET mechanism. One can of course argue that there is always a risk for aggregation between AuNPs induced by the multivalency of streptavidin (4 binding pockets per molecule). This might occur when there is more than one peptide per AuNP (as for the 6 and 18 nm particles). However, no colour change nor any plasmon band shift were observed after mixing SA488 with the 6 nm AuNPs at the molar ratio of AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 of 3
SA488 of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, indicating negligible aggregation of the AuNPs (Fig. S4†).
1, indicating negligible aggregation of the AuNPs (Fig. S4†).
AuNPs size-dependent protease activity
The BoLcA activity was monitored through time-dependent fluorescence spectra of SA488 incubated with 1.4 nm AuNP–peps which were pre-treated with BoLcA for 30 min to 6 hours (Fig. 3A). The fluorescence intensity at the emission wavelength of 520 nm increased from 2.2 × 105 to 3.2 × 105 cps μA−1 after a 0.5 h incubation with 10 nM BoLcA and reached saturation at 4.0 × 105 cps μA−1 after a 2 h incubation.The time-dependent peptide cleavage for AuNPs with diameters of 1.4 nm, 6 nm and 18 nm is demonstrated in Fig. 3B. The kinetics show saturation in 2 to 5 hours for the 1.4 nm AuNP–pep (200 nM) after exposure to 3 nM and 10 nM BoLcA, with an initial catalytic rate v = 0.6 nM min−1 and 2 nM min−1, respectively. This corresponds to an initial turnover number of k = 0.2 min−1. The corresponding initial turnover number for 6 nm and 18 nm AuNPs is 0.0025 min−1, which is 80-fold lower than that for the 1.4 nm AuNPs. The k values were calculated based on a coverage of 3 peptides per 6 nm AuNP (12 nM) and 51 peptides per 18 nm AuNP (1 nM) which were cleaved initially at a rate of 0.78 and 0.75 peptides per hour with 5 nM BoLcA, respectively. The rate constant is calculated as KT = kcat/KM and equals 2.8 × 106 M−1 min−1 (see Fig. 3D and details in ESI†), based on the fitting of the kinetics for 1.4 nm AuNP–pep incubation with 3 nM and 10 nM BoLcA. Assuming kcat = 30 s−1 as reported with the same peptide sequence,21 the Michaelis–Menten constant KM is estimated to equal KM = 643 μM, which is comparable to that of the free peptide.21 The poorer activity of BoLcA for the peptide on the large AuNPs is ascribed to the fact that the active pocket of BoLcA is deeply buried inside the enzyme (∼2.4 nm),26 and steric hindrance induced by the AuNPs.
A similar phenomenon was also observed for trypsin and the same AuNP–pep. The catalytic rate of trypsin was higher for small AuNPs than for large AuNPs as indicated in Fig. 3C. The cleavage percentage was defined as ΔF/(FSA488 − F0), where FSA488 and F0 are the fluorescence intensities of free SA488 and AuNP–pep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SA488 conjugates measured at λ = 520 nm, respectively. The results indicated that about 90% of peptides on AuNPs were hydrolyzed by trypsin after 1, 4 and 10 hours for 1.4 nm, 6 nm and 18 nm AuNP–pep, respectively. As compared with BoLcA (Fig. 3B), trypsin hydrolyzed a higher amount of peptide in a shorter reaction time, indicating a higher activity. The rate constant KT was estimated as KT = 8.1 × 107 M−1 min−1, 7.2 × 106 M−1 min−1 and 4.42 × 106 M−1 min−1 for the AuNPs with diameters of 1.4 nm, 6 nm and 18 nm, respectively (Fig. 3D). The KT values are comparable to previously reported values 7.8 × 107–2.34 × 108 M−1 min−1 for the digestion of a fifteen-residue peptide substrate by trypsin,27 but 3 orders of magnitude higher than that reported for four-residue peptide substrates containing an arginine cleavage site.28 The trypsin proteolytic activity is about 29 times higher than that of BoLcA. Furthermore, upon increasing the size of AuNPs from 1.4 nm to 18 nm in diameter, the trypsin activity decreased by 18 fold, a decrease that was 4 times smaller than the 80-fold decrease for BoLcA (Fig. 3D). Again the poor catalytic activity of BoLcA and more significant hindrance by large AuNPs might be ascribed to the deeper active pocket sites and 2-fold higher molecular weight of BoLcA as compared to trypsin, as well as the smaller number of cleavage sites provided by the peptide substrate. There are 4 trypsin cleavage sites in total (i.e. lysine K and arginine R) and only one (QR) for BoLcA.
SA488 conjugates measured at λ = 520 nm, respectively. The results indicated that about 90% of peptides on AuNPs were hydrolyzed by trypsin after 1, 4 and 10 hours for 1.4 nm, 6 nm and 18 nm AuNP–pep, respectively. As compared with BoLcA (Fig. 3B), trypsin hydrolyzed a higher amount of peptide in a shorter reaction time, indicating a higher activity. The rate constant KT was estimated as KT = 8.1 × 107 M−1 min−1, 7.2 × 106 M−1 min−1 and 4.42 × 106 M−1 min−1 for the AuNPs with diameters of 1.4 nm, 6 nm and 18 nm, respectively (Fig. 3D). The KT values are comparable to previously reported values 7.8 × 107–2.34 × 108 M−1 min−1 for the digestion of a fifteen-residue peptide substrate by trypsin,27 but 3 orders of magnitude higher than that reported for four-residue peptide substrates containing an arginine cleavage site.28 The trypsin proteolytic activity is about 29 times higher than that of BoLcA. Furthermore, upon increasing the size of AuNPs from 1.4 nm to 18 nm in diameter, the trypsin activity decreased by 18 fold, a decrease that was 4 times smaller than the 80-fold decrease for BoLcA (Fig. 3D). Again the poor catalytic activity of BoLcA and more significant hindrance by large AuNPs might be ascribed to the deeper active pocket sites and 2-fold higher molecular weight of BoLcA as compared to trypsin, as well as the smaller number of cleavage sites provided by the peptide substrate. There are 4 trypsin cleavage sites in total (i.e. lysine K and arginine R) and only one (QR) for BoLcA.
BoLcA sensing
Considering the situation that the large AuNPs significantly hindered the accessibility of BoLcA to the peptides on the surface, we employed the smallest AuNPs (i.e. 1.4 nm in diameter) for the detection of BoLcA. The time-dependent fluorescence intensity of SA488 was monitored after incubation with AuNP–pep which was pre-treated with BoLcA at various concentrations from 1 pM to 10 nM at 37 °C (Fig. 4A). The fluorescence intensity saturated quickly after 2 hours incubation of AuNP–pep with BoLcA at concentrations higher than 3 nM (Fig. 4A). However, at low concentrations of BoLcA (<3 nM), the fluorescence intensity increased at an approximately constant rate after 2 hours of incubation. It is interesting to note that at concentrations of BoLcA lower than 3 nM the fluorescence intensity decreased in the first reaction hour (see insert in Fig. 4A). This might tentatively be ascribed to a transient conformational change of the peptide upon favourable interaction with BoLcA prior to cleavage. This hypothesis is based on the assumption that the fluorescence decline is due to a decrease in the distance between the dye and the AuNP. The same trend was not observed for trypsin at concentrations down to 1 pM because of its higher catalytic rate and its less deep catalytic/cleavage site compared to BoLcA (Fig. 3D and 4A). However, to confirm whether the peptide changes conformation upon affinity interaction with BoLcA will require more advanced single-molecule measurements,29 experiments that are beyond the scope of the present study.The calibration curve for the detection of BoLcA from 1 pM to 10 nM reveals a limit of detection (LOD) of 1 pM (50 pg ml−1), which is determined as the concentration of BoLcA at which the fluorescence intensity change ΔF is three times the standard deviation of the fluorescence fluctuation of the control samples, i.e. 3 × ΔF = 1000 cps μA−1. The method enabled the sensitive detection of BoLcA with an assay time of 2–3 hours which is comparable to the vesicle amplified SPR sandwich assay (LOD ∼0.3 to 10 pM and assay time of 10 min to several hours),10 aptamer-based electrochemical assay (LOD of 40 pg ml−1 and assay time of 24 h),30 and the antibody-based SPR sandwich assay for the detection of BoNT Type B in buffer and honey (LOD ∼ 2 pM and assay time of 1–2 h).7 The LOD is about 2 to 3 orders of magnitude lower than the quantum dots-based FRET method (LOD ∼ 350 pM and assay time of 2–3 h),31 antibody-based ELISA (LOD of 0.2–2 ng ml−1 and assay time of 8 h).5 But it is less sensitive than mouse lethality assay (LOD of 20–30 pg ml−1 and assay time of 2–4 days).32 However, our method typically provides a shorter assay time than that of conventional ELISA and mouse lethality assays, and is expected to be a more robust assay than the antibody-based assays.
Conclusion
We have designed and synthesized a substrate peptide, which was immobilized on AuNPs with diameters of 1.4–18 nm, for the detection of BoLcA activity using a fluorescence energy transfer assay format. The results indicated that larger AuNPs provided higher quenching efficiencies, but significantly hindered the accessibility of BoLcA to the peptides on the AuNP surface, thus leading to an 80-fold lower initial catalytic rate for BoLcA. The assay exhibited a limit of detection of 1 pM for the detection of BoLcA with an assay time of 2–3 hours which is faster and more sensitive than many other assays including conventional ELISA. The proposed assay, based on a synthetic peptide, is also more robust than antibody-based assays.Acknowledgements
This research is supported by the Science & Engineering Research Council (SERC) of the Agency for Science, Technology and Research (A*STAR), for projects under the number 1021520015.References
- C. Montecucco and J. Molgo, Curr. Opin. Pharmacol., 2005, 5, 274–279 CrossRef CAS PubMed.
- (a) B. R. Singh, Neurotoxic. Res., 2006, 9, 73–92 CrossRef CAS; (b) G. Schiavo, M. Matteoli and C. Montecucco, Physiol. Rev., 2000, 80, 717–766 CAS; (c) L. Li and B. R. Singh, J. Toxicol., Toxin Rev., 1999, 18, 95–112 CrossRef CAS.
- S. S. Arnon, R. Schechter, T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, J. Hauer, M. Layton, S. Lillibridge, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, D. L. Swerdlow, K. Tonat and W. G. C. Biodefense, JAMA, J. Am. Med. Assoc., 2001, 285, 1059–1070 CrossRef CAS.
- S. K. Sharma and R. C. Whiting, J. Food Prot., 2005, 68, 1256–1263 CAS.
- (a) M. A. Poli, V. R. Rivera and D. Neal, Toxicon, 2002, 40, 797–802 CrossRef CAS; (b) M. Szilagyi, V. R. Rivera, D. Neal, G. A. Merrill and M. A. Poli, Toxicon, 2000, 38, 381–389 CrossRef CAS.
- (a) J. T. Mason, L. X. Xu, Z. M. Sheng and T. J. O'Leary, Nat. Biotechnol., 2006, 24, 555–557 CrossRef CAS PubMed; (b) J. T. Mason, L. Xu, Z. M. Sheng, J. He and T. J. O'Leary, Nat. Protoc., 2006, 1, 2003–2011 CrossRef CAS PubMed; (c) H. C. Wu, Y. L. Huang, S. C. Lai, Y. Y. Huang and M. F. Shalo, Lett. Appl. Microbiol., 2001, 32, 321–325 CrossRef CAS.
- J. Ladd, A. D. Taylor, J. Homola and S. Y. Jiang, Sens. Actuators, B, 2008, 130, 129–134 CrossRef CAS PubMed.
- (a) V. R. Rivera, F. J. Gamez, W. K. Keener, J. A. White and M. A. Poli, Anal. Biochem., 2006, 353, 248–256 CrossRef CAS PubMed; (b) V. Guglielmo-Viret, O. Attree, V. Blanco-Gros and P. Thullier, J. Immunol. Methods, 2005, 301, 164–172 CrossRef CAS PubMed.
- J. W. Grate, R. M. Ozanich, M. G. Warner, J. D. Marks and C. J. Bruckner-Lea, TrAC, Trends Anal. Chem., 2010, 29, 1137–1156 CrossRef CAS PubMed.
- G. Ferracci, S. Marconi, C. Mazuet, E. Jover, M. P. Blanchard, M. Seagar, M. Popoff and C. Leveque, Anal. Biochem., 2011, 410, 281–288 CrossRef CAS PubMed.
- P. Capek, K. S. Kirkconnell and T. J. Dickerson, J. Am. Chem. Soc., 2010, 132, 13126–13128 CrossRef CAS PubMed.
- (a) S. Sun, J. Francis, K. E. Sapsford, Y. Kostov and A. Rasooly, Sens. Actuators, B, 2010, 146, 297–306 CrossRef CAS PubMed; (b) D. Min, W. H. Tepp, E. A. Johnson and E. R. Chapman, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14701–14706 CrossRef PubMed; (c) J. J. Schmidt, R. G. Stafford and C. B. Millard, Anal. Biochem., 2001, 296, 130–137 CrossRef CAS PubMed; (d) J. A. Ross, M. A. Gilmore, D. Williams, K. R. Aoki, L. E. Steward and D. M. Jameson, Anal. Biochem., 2011, 413, 43–49 CrossRef CAS PubMed.
- (a) Y. Choi, J. Lee, K. Kim, H. Kim, P. Sommer and R. Song, Chem. Commun., 2010, 46, 9146–9148 RSC; (b) Y. H. Wang, P. Shen, C. Y. Li, Y. Y. Wang and Z. H. Liu, Anal. Chem., 2012, 84, 1466–1473 CrossRef CAS PubMed; (c) T. Zauner, R. Berger-Hoffmann, K. Muller, R. Hoffmann and T. Zuchner, Anal. Chem., 2011, 83, 7356–7363 CrossRef CAS PubMed; (d) J. Karvinen, V. Laitala, M. L. Makinen, O. Mulari, J. Tamminen, J. Hermonen, P. Hurskainen and I. Hemmila, Anal. Chem., 2004, 76, 1429–1436 CrossRef CAS PubMed; (e) D. E. Prasuhn, A. Feltz, J. B. Blanco-Canosa, K. Susumu, M. H. Stewart, B. C. Mei, A. V. Yakovlev, C. Loukov, J. M. Mallet, M. Oheim, P. E. Dawson and I. L. Medintz, ACS Nano, 2010, 4, 5487–5497 CrossRef CAS PubMed.
- (a) I. L. Medintz, A. R. Clapp, F. M. Brunel, T. Tiefenbrunn, H. T. Uyeda, E. L. Chang, J. R. Deschamps, P. E. Dawson and H. Mattoussi, Nat. Mater., 2006, 5, 581–589 CrossRef CAS PubMed; (b) K. Boeneman, B. C. Mei, A. M. Dennis, G. Bao, J. R. Deschamps, H. Mattoussi and I. L. Medintz, J. Am. Chem. Soc., 2009, 131, 3828–3829 CrossRef CAS PubMed; (c) H. Q. Yao, Y. Zhang, F. Xiao, Z. Y. Xia and J. H. Rao, Angew. Chem., Int. Ed., 2007, 46, 4346–4349 CrossRef CAS PubMed; (d) L. F. Shi, V. De Paoli, N. Rosenzweig and Z. Rosenzweig, J. Am. Chem. Soc., 2006, 128, 10378–10379 CrossRef CAS PubMed; (e) W. R. Algar, A. Malonoski, J. R. Deschamps, J. B. Banco-Canosa, K. Susumu, M. H. Stewart, B. J. Johnson, P. E. Dawson and I. L. Medintz, Nano Lett., 2012, 12, 3793–3802 CrossRef CAS PubMed.
- (a) R. Hardman, Environ. Health Perspect., 2006, 114, 165–172 CrossRef; (b) M. Bottrill and M. Green, Chem. Commun., 2011, 47, 7039–7050 RSC; (c) L. Y. T. Chou and W. C. W. Chan, Nat. Nanotechnol., 2012, 7, 416–417 CrossRef CAS PubMed.
- (a) M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2010, 132, 9383–9391 CrossRef CAS PubMed; (b) T. L. Jennings, M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2006, 128, 5462–5467 CrossRef CAS PubMed; (c) J. R. Lakowicz, Anal. Biochem., 2005, 337, 171–194 CrossRef CAS PubMed; (d) T. Pons, I. L. Medintz, K. E. Sapsford, S. Higashiya, A. F. Grimes, D. S. English and H. Mattoussi, Nano Lett., 2007, 7, 3157–3164 CrossRef CAS PubMed; (e) Y. Wang, L. Wu, X. D. Zhou, T. I. Wong, J. L. Zhang, P. Bai, E. P. Li and B. Liedberg, Sens. Actuators, B, 2013, 186, 205–211 CrossRef CAS PubMed.
- (a) B. Dubertret, M. Calame and A. J. Libchaber, Nat. Biotechnol., 2001, 19, 365–370 CrossRef CAS PubMed; (b) H. Wang, J. S. Li, Y. X. Wang, J. Y. Jin, R. H. Yang, K. M. Wang and W. H. Tan, Anal. Chem., 2010, 82, 7684–7690 CrossRef CAS PubMed; (c) F. Li, H. Pei, L. H. Wang, J. X. Lu, J. M. Gao, B. W. Jiang, X. C. Zhao and C. H. Fan, Adv. Funct. Mater., 2013, 23, 4140–4148 CrossRef CAS.
- (a) D. S. Seferos, D. A. Giljohann, H. D. Hill, A. E. Prigodich and C. A. Mirkin, J. Am. Chem. Soc., 2007, 129, 15477–15479 CrossRef CAS PubMed; (b) J. P. Xue, L. L. Shan, H. Y. Chen, Y. Li, H. Y. Zhu, D. W. Deng, Z. Y. Qian, S. Achilefu and Y. Q. Gu, Biosens. Bioelectron., 2013, 41, 71–77 CrossRef CAS PubMed; (c) N. Li, C. Y. Chang, W. Pan and B. Tang, Angew. Chem., Int. Ed., 2012, 51, 7426–7430 CrossRef CAS PubMed.
- (a) Y. P. Kim, Y. H. Oh, E. Oh, S. Ko, M. K. Han and H. S. Kim, Anal. Chem., 2008, 80, 4634–4641 CrossRef CAS PubMed; (b) D. W. Deng, D. Y. Zhang, Y. Li, S. Achilefu and Y. Q. Gu, Biosens. Bioelectron., 2013, 49, 216–221 CrossRef CAS PubMed.
- (a) D. Zheng, D. S. Seferos, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 3258–3261 CrossRef CAS PubMed; (b) X. Y. Huang, T. Lan, B. C. Zhang and J. C. Ren, Analyst, 2012, 137, 3659–3666 RSC.
- J. J. Schmidt and K. A. Bostian, J. Protein Chem., 1997, 16, 19–26 CrossRef CAS.
- X. Liu, Y. Wang, P. Chen, Y. Wang, J. Zhang, D. Aili and B. Liedberg, Anal. Chem., 2014, 86, 2345–2352 CrossRef CAS PubMed.
- J. J. Schmidt and K. A. Bostian, J. Protein Chem., 1995, 14, 703–708 CrossRef CAS.
- P. Chen, R. Selegard, D. Aili and B. Liedberg, Nanoscale, 2013, 5, 8973–8976 RSC.
- S. Link and M. A. El-Sayed, Int. Rev. Phys. Chem., 2000, 19, 409–453 CrossRef CAS.
- R. Kukreja and B. R. Singh, in Microbial Toxins: Current Research and Future Trends, Caister Academic Press, 2009 Search PubMed.
- G. S. Coombs, M. S. Rao, A. J. Olson, P. E. Dawson and E. L. Madison, J. Biol. Chem., 1999, 274, 24074–24079 CrossRef CAS PubMed.
- R. M. Caprioli and L. Smith, Anal. Chem., 1986, 58, 1080–1083 CrossRef CAS.
- X. Michalet, S. Weiss and M. Jager, Chem. Rev., 2006, 106, 1785–1813 CrossRef CAS PubMed.
- F. Wei and C. M. Ho, Anal. Bioanal. Chem., 2009, 393, 1943–1948 CrossRef CAS PubMed.
- K. E. Sapsford, J. Granek, J. R. Deschamps, K. Boeneman, J. B. Blanco-Canosa, P. E. Dawson, K. Susumu, M. H. Stewart and I. L. Medintz, ACS Nano, 2011, 5, 2687–2699 CrossRef CAS PubMed.
- M. Lindstrom and H. Korkeala, Clin. Microbiol. Rev., 2006, 19, 298–314 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of experiments, FRET calculation, MALDI-TOF, DLS characterization and peptide-ratio dependent quenching efficiency of AuNPs. See DOI: 10.1039/c3sc53305k |
| This journal is © The Royal Society of Chemistry 2014 |