Open Access Article
Open Access ArticleStimuli-induced folding cascade of a linear oligomeric guest chain programmed through cucurbit[n]uril self-sorting (n = 6, 7, 8)†
Luca
Cera
and
Christoph A.
Schalley
*
Institut für Chemie und Biochemie, Freie Universität Berlin, Takustrasse 3, 14195 Berlin, Germany. E-mail: christoph@schalley-lab.de
First published on 14th April 2014
Abstract
A six-station linear guest for cucurbit[7]uril and cucurbit[8]uril has been synthesized in order to implement a cascade of transformations driven by external stimuli. The guest chain is sequence-programmed with electron-deficient viologen and electron-rich naphthalene stations linked by either flexible or rigid spacers that affect the chain's folding properties. Together with the orthogonal guest selectivity of the two cucurbiturils, these properties result in self-sorted cucurbituril pseudorotaxane foldamers. Each transformation is controlled by suitable chemical and redox inputs and leads not only to refolding of the guest chain, but also to the liberation of secondary messenger molecules which render the system presented here reminiscent of natural signaling cascades. The steps of the cascade are analyzed by UV/Vis, 1H NMR and electrospray (tandem) mass spectrometry to investigate the different pseudorotaxane structures in detail. With one guest oligomer, three different cucurbiturils, and several different chemical and redox inputs, a chemical system is created which exhibits complex behavior beyond the chemist's paradigm of the pure chemical compound.
Introduction
Systems chemistry is a new emerging field in chemistry1 which breaks with the current paradigm of the pure, isolated compound which has been one of the foundations of synthetic chemistry for decades. In contrast, systems chemistry aims at complex mixtures of compounds which cooperate synergistically in reactivity networks. The metabolism of a living cell offers the role model for such a chemical network. It does not only produce and metabolize the compounds needed for homeostasis and reproduction, but also regulates itself in spatiotemporal patterns of utmost complexity far away from thermodynamic equilibrium. Regulation often involves signal transduction through membranes so that outer stimuli can affect the living cell to react to its environment. But also inside a cell, signaling cascades operate in which enzymes refold in conformation upon a messenger binding to a certain receptor site and thus inducing an active conformation that generates secondary messengers.A concise description of such a network would require the detailed knowledge of the differential equations describing the concentration changes for all metabolites involved. Even then, the pattern evolution can only be modeled numerically. Feedback loops remain problematic; in particular as self-accelerating processes2 tend to amplify small initial deviations from a given set of starting parameters put into such a calculation. As a major consequence, predicting the long-term behavior of a complex chemical system is at least difficult, if not impossible. The more so, this is true for the de novo design of synthetic chemical networks with a certain desired behavior.
Another interesting conceptual idea evolves, if one accepts that there is no fundamental difference between living and inanimate matter: evolution is then not restricted to the biological world anymore, but can become an emergent property3 of a synthetic system as well.4 Consequently, the motivation to investigate chemical networks comes not only from the desire to emulate and understand nature,5 but also from the interest in creating chemical systems to put our conceptions on evolution and emergence to the test – or to create systems that have so far new and unseen properties.
Several different approaches have led to systems displaying emergent properties, for example self-sorting in supramolecular systems,6 the adaptive behavior of dynamic combinatorial libraries (DCLs),7 self-replicators that feed on DCLs,8 or mechanosensitive supramolecular fiber formation.9 Recently, Nitschke et al. reported metallo-supramolecular systems which are responsive to external stimuli and undergo cascading transformations that are reminiscent of signaling cascades in living cells.10
During the last decades, the host–guest chemistry of the cucurbit[n]uril family11 (n = 5–8, 10) has seen tremendous interest mainly because of strong binding constants,12 a wide variety of guests, the cavity-size-dependent differences in guest preferences,13 and the potential to implement switching behavior.14 Only recently, few groups have been focusing on the dynamic self-sorting of these hosts.15 Previously, we reported a detailed study on the social self-sorting of cucurbit[7]uril (CB7) and cucurbit[8]uril (CB8) in the presence of guests like viologen, dihydroxy naphthalene and the corresponding covalently linked di- and trimers of both.16 As a result, specific supramolecular architectures form in this social network, because CB7 binds viologen dications, while CB8 prefers guest pairs of electron-deficient viologen and electron-rich dihydroxy naphthalene.17 One of the still quite rare cases has been discovered, in which both hosts CB7 and CB8 have a preference for the same guest A over B, but still socially self-sort when stoichiometry is controlled, because the binding constants of the CB7·A and CB8·A pairs are sufficiently different. Besides pairs of electron-deficient and electron-rich guests, CB8 also strongly binds dimers of viologen cation-radicals.18 This motif has found applications in the design of redox-stimulus-driven molecular machines,18 but no studies are available that discuss how the cation-radical dimer affects the self-sorting behavior of CB7 and CB8. Consequently, cucurbiturils have large potential for studying their behavior in complex chemical systems.
Here, we present the five-step transformation cascade shown in Fig. 1. The cascade involves a linear guest chain BATCl8 with six stations which allow inducing the formation of different foldamers by different signals added to the guest. Elements of the underlying program include the different cavity sizes and thus different guest selectivities of the three different cucurbit[n]urils CB6–CB8 (n = 6–8), the sequence of stations along the guest chain and the rigidity/flexibility of the spacers connecting them. This transformation cascade is thus reminiscent of nature's signaling cascades in that external signals cause recognition events that lead to controlled conformational changes in a sequence-programmed chain and result in the liberation of secondary messenger molecules.
Results and discussion
Concept and guest chain synthesis
To implement the stimuli-induced folding processes depicted in Fig. 1, the guest molecule BATCl8 is designed to have a specific sequence of guest units: two terminal viologens (T) are connected to the adjacent naphthalenes through flexible propyleneoxy linkers; the two inner viologens (I) are linked to the naphthalenes through more rigid triazolylethylene spacers. The central connection between the two inner viologens is again a flexible propylene group. The propyleneoxy linkers are known to be the shortest linkers permitting the viologen–naphthalene guest dimer to fold into a charge-transfer (CT) complex of the electron-poor viologen and the electron-rich naphthalene units.19 The central propylene linker instead helps maximizing the formation of the cofacial viologen cation-radical dimer upon two one-electron reductions at the central viologen moieties.20 In contrast, suitable folding into CT complexes of the central viologen and the naphthalene units is made impossible by the more rigid triazolylethylene spacers. Thus, the guest chain involves two design elements: the sequence of binding stations and the incorporation of suitable spacers, which either allow or prohibit folding of the chain, respectively.BATCl8 is synthesized in a convergent manner by initially connecting the two terminal viologen–naphthalene pairs (see ESI† for details). For solubility, the azide-substituted hydroxynaphthalene is first equipped with a bromopropylene side chain and then reacted with N-methyl-4,4′-bipyridinium iodide. For the synthesis of the inner part of the guest, 4-bromo-1-butyne is reacted with 4,4′-bipyridine. Before connecting the two inner viologens to each other, an anion exchange of the bromide to the corresponding hexafluorophosphate is necessary for solubility reasons. After this exchange, the two inner viologens are connected with 1,3-dibromopropane and the product subsequently equipped with the two terminal parts through a copper-catalyzed Huisgen–Sharpless–Meldal click reaction. In order to obtain the final, water-soluble chloride salt, another anion exchange is performed.
Characterization of the cascade intermediates
With each step of the transformation cascade, new external stimuli are added either as chemical or redox signals. Consequently, the mixture becomes more and more complex in each step. Classical NMR analysis of complexation-induced signal shifts can provide very useful structural information on the host–guest complexes formed. However, the interpretation of the NMR spectra might become increasingly difficult with the complexity of the mixture under study. A second complementary method is thus advantageous to cross check the results obtained from NMR experiments.The BAT8+ guest chain is – as well as the other guests DAH2+ and ADA+ – charged and thus facilitates a mass-spectrometric analysis of the mixtures obtained after each step in the cascade. This method has been utilized before and showed that even different isomeric host–guest complexes can be distinguished by their fragmentation patterns.16 Therefore, (tandem) electrospray Fourier-transform ion-cyclotron-resonance mass spectrometry (ESI-FTICR-MS) is used in the present study as one of the major methods for the characterization of the different host–guest complexes formed in each of the transformation steps along the cascade. There are three peculiarities in the gas-phase behaviour of viologens and their cucurbituril complexes, which are important for understanding the corresponding mass spectra: (i) the naked viologen dications are prone to fragmentation induced by charge repulsion unless stabilized by host–guest interactions or the presence of counterions.21 (ii) When cucurbiturils are bound to the viologens, most often the highest possible charge states are observed with remarkably high intensity. This is in contrast to the free guests, which usually appear in lower charge states because they carry a number of counterions.16 This finding can be rationalized by the increase in anion–cation distance during complexation of the cucurbituril. It is thus by far easier to strip off the counterions from the cucurbituril complexes than from the free guests upon ionization. Nevertheless, charge-state distributions are often observed. (iii) Viologen dications and cation-radicals sometimes undergo one-electron reduction or oxidation reactions at the ESI capillary so that charge states can be observed other than those expected from the number of counterions still present in the complexes.
In addition, UV/Vis spectroscopy provides insight into the formation of the viologen cation-radicals and their dimerization products upon reduction of BATCl8. These experiments are thus helpful, when NMR experiments are hampered by the paramagnetism of the viologen cation-radicals.
First step: complex formation of BATCl8 with CB7
The first step of the transformation cascade is the formation of a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
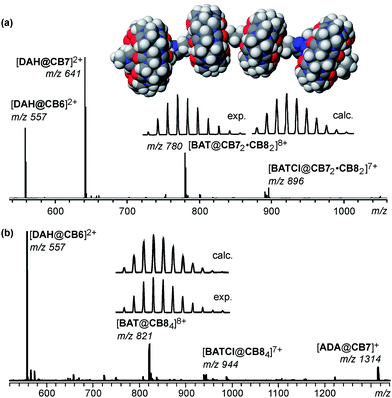
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex of CB7 and BATCl8. The binding constant of methyl viologen to CB7 has been reported22 to amount to K = 2.0 × 105M−1, a value certainly high enough to justify the expectation that this complex will form. It is simply generated by adding 4 eq. of CB7 to a 0.857 mM water solution of BATCl8. After equilibration, the expected host–guest complex [BAT@CB74]8+ is detected as the base peak in the electrospray Fourier-transform ion-cyclotron-resonance (ESI-FTICR) mass spectra at m/z 738 (Fig. 2, center). In addition, the [BATCl@CB74]7+ (m/z 849) and [BATCl2@CB74]6+ (m/z 996) ions are observed with lower intensities. Also traces of [BAT@CB75]8+ (m/z 884) are detected, which is likely an unspecific complex as often detected in ESI mass spectra. This assignment is supported by a fragmentation experiment (see ESI†). Only those fragments are observed that are also found for [BAT@CB74]8+. Consequently, the fifth CB7 is only very weakly bound and easily stripped off the complex. Finally, the [BAT@CB73]8+ ion at m/z 593 is generated by loss of a CB7 from [BAT@CB74]8+. One viologen is not stabilized by the cucurbituril anymore and thus a charge separating fragmentation into the fragments [A@CB7]3+ (m/z 528) and [B@CB72]5+ (m/z 626) occurs easily. Consequently, this mass spectrum is in good agreement with expectation for the formation of [BAT@CB74]8+.
1 complex of CB7 and BATCl8. The binding constant of methyl viologen to CB7 has been reported22 to amount to K = 2.0 × 105M−1, a value certainly high enough to justify the expectation that this complex will form. It is simply generated by adding 4 eq. of CB7 to a 0.857 mM water solution of BATCl8. After equilibration, the expected host–guest complex [BAT@CB74]8+ is detected as the base peak in the electrospray Fourier-transform ion-cyclotron-resonance (ESI-FTICR) mass spectra at m/z 738 (Fig. 2, center). In addition, the [BATCl@CB74]7+ (m/z 849) and [BATCl2@CB74]6+ (m/z 996) ions are observed with lower intensities. Also traces of [BAT@CB75]8+ (m/z 884) are detected, which is likely an unspecific complex as often detected in ESI mass spectra. This assignment is supported by a fragmentation experiment (see ESI†). Only those fragments are observed that are also found for [BAT@CB74]8+. Consequently, the fifth CB7 is only very weakly bound and easily stripped off the complex. Finally, the [BAT@CB73]8+ ion at m/z 593 is generated by loss of a CB7 from [BAT@CB74]8+. One viologen is not stabilized by the cucurbituril anymore and thus a charge separating fragmentation into the fragments [A@CB7]3+ (m/z 528) and [B@CB72]5+ (m/z 626) occurs easily. Consequently, this mass spectrum is in good agreement with expectation for the formation of [BAT@CB74]8+.
 | ||
| Fig. 2 Top: force-field optimized structure (MM2, CaChe 6.1 program, Fujitsu, Krakow, Poland) of the [BAT@CB74]8+ complex. Center: electrospray FTICR mass spectrum of the 0.857 mM sample solution of BATCl8 and 4 eq. of CB7 in water (after dilution to ca. 30 μM). Bottom: infrared multiphoton dissociation experiment with mass-selected [BAT@CB74]8+ ions. Also, see ESI† for details. | ||
When mass-selected [BAT@CB74]8+ ions are fragmented with a CO2 IR laser in infrared multiphoton dissociation (IRMPD) experiments (Fig. 2, bottom), three competing pathways are observed in a rather complex fragmentation behavior. The loss of one CB7 with subsequent dissociation into [A@CB7]3+ and [B@CB72]5+ is the first and likely least energy-demanding one of them. The second fragmentation channel is a charge-separating dissociation into fragments [A@CB7]3+ and [C@CB73]5+ (m/z 859). The third reaction is cleavage of the parent ion into [B@CB72]5+ and [A@CB72]3+ (m/z 916). The [C@CB73]5+ ion undergoes CB7 and nitrogen molecule23 losses followed by a fragmentation into [A@CB7]3+ and [D@CB7]2+ (m/z 758). Finally, [E@CB7]2+ (m/z 667) is the likely end point of several fragmentation pathways.
This analysis of the reaction pathways is based on the experience that a multiply charged ion preferentially fragments into two smaller daughter ions, which are both charged and thus should both appear in the spectrum. With this prerequisite, a number of other potential channels can safely be ruled out as only one of the expected product ions is observed. The fact that cucurbituril loss competes with covalent bond cleavages is indicative of the formation of rather strong supramolecular host–guest interactions and thus points to the formation of a specific 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex.
1 complex.
The 1H and 1H,1H COSY NMR spectra (see ESI†) of this complex reveal the two outer cucurbiturils to reside almost centrally on the terminal viologens. However, the CB7 molecules bound to the inner viologens appear with two sets of signals. The less intense one corresponds to CB7 located centrally on the inner viologens. The more intense one can be assigned to a CB7 that is shifted towards the triazole ring. As this structure is the major species in the equilibrium, it is likely that the triazole C–H hydrogen forms a C–H⋯O hydrogen bond24 with one of the carbonyl groups in the CB7 outer rim as also indicated by the significant downfield shift of the triazole C–H signal (Δδ ≈ 0.5 ppm). Similar “half-threaded” complexes have been observed earlier.25 Consequently, the NMR spectra are in line with the formation of a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex and add more structural information in that they show isomeric complexes to exist in an equilibrium that is on slow exchange with respect to the NMR time scale.
1 complex and add more structural information in that they show isomeric complexes to exist in an equilibrium that is on slow exchange with respect to the NMR time scale.
Second step: competition between CB7 and CB8
In the second step of the transformation cascade, 2 eq. of CB8 are added in competition to the already present CB7 hosts. CB8 binds methyl viologen with a binding constant similar to that of CB7 (K = 1.1 × 105M−1).26 With its larger cavity, it can also very favorably bind a charge-transfer guest pair. For example, the binding constant of the methyl viologen–dihydroxynaphthalene pair amounts to K = K1 × K2 ≈ 109M−2.27 Earlier studies16 revealed a clear self-sorting phenomenon: when an electron-deficient viologen and an electron-rich naphthalene are connected covalently, CB8 binds significantly stronger to this pair than a CB7 to the viologen alone. Based on this self-sorting behavior, the addition of CB8 to the sample solution obtained in the first step should lead to the formation of the BATCl8@CB72·CB82 complex with a well-defined CB8-CB7-CB7-CB8 sequence. The external CB8 signal is thus expected to give rise to a refolding of the guest chain together with the liberation of two molecules of CB7.Clear-cut evidence for this successful transformation comes from ESI mass spectrometry as well as NMR spectroscopic experiments (Fig. 3). The ESI-FTICR mass spectrum of the sample after CB8 addition exhibits only one major signal which can be assigned to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex [BAT@CB72·CB82]8+ (m/z 780). Again, smaller signals for the next two lower charge states are observed as well as two unspecific complexes, which carry either a CB7 or a CB8 in excess. So far, the spectrum is analogous to that of the [BAT@CB74]8+ complex discussed above. However, one remarkable difference is obvious: there are no fragments formed that could be traced back to the loss of a cucurbituril. This can be attributed to the stronger binding of the two CB8 molecules, which act as stoppers trapping the two inner CB7 molecules. This finding already provides a first indication of the cucurbituril positions along the chain.
2 complex [BAT@CB72·CB82]8+ (m/z 780). Again, smaller signals for the next two lower charge states are observed as well as two unspecific complexes, which carry either a CB7 or a CB8 in excess. So far, the spectrum is analogous to that of the [BAT@CB74]8+ complex discussed above. However, one remarkable difference is obvious: there are no fragments formed that could be traced back to the loss of a cucurbituril. This can be attributed to the stronger binding of the two CB8 molecules, which act as stoppers trapping the two inner CB7 molecules. This finding already provides a first indication of the cucurbituril positions along the chain.
These considerations are supported by the IRMPD experiment (Fig. 3b) performed with mass-selected [BAT@CB72·CB82]8+. In contrast to that done with [BAT@CB74]8+, only a single fragmentation pathway is observed – a charge separating cleavage into [A@CB8]3+ (m/z 584) and [C@CB72·CB8]2+ (m/z 892). No loss of CB8 competes with this covalent bond cleavage reaction. This points again to the significantly enhanced binding strength of CB8. More importantly, no [A@CB7]3+ fragment ion is observed thus providing evidence for the relative positioning of CB7 in the center and CB8 at the termini of the guest chain.
A comparison of the 1H NMR spectra of the free host BATCl8 and the BATCl8@CB72·CB82 complex (Fig. 3c and d) reveals the expected complexation-induced shifts. All viologen β protons reside inside hydrophobic cavities and experience upfield shifts of about 1.5 ppm.22 The same holds true for the naphthalene protons so that we can safely conclude the naphthalene units to be located inside the cavities of the two CB8 molecules. The viologen α protons do not move as much as they are located at the rims of the cucurbiturils. While the αI protons for the inner viologens experience a minor downfield shift, the αT protons of the terminal viologens are shifted somewhat upfield indicating that they are affected by the anisotropy of the adjacent naphthalene. Consequently, the NMR shifts are structure-indicative and confirm the terminally folded 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes of BATCl8 and the two cucurbiturils. Again, the triazole C–H signal is shifted downfield indicating that C–H⋯O hydrogen bonds are also formed in this complex. In contrast to BATCl8@CB74, however, no positionally shifted isomers are observed anymore. We conclude the CB8 host molecules to bind to the triazole C–H with one of their carbonyl oxygen atoms. Therefore, the CB7 molecules now prefer to be located centrally on the inner viologens and do not shift towards the triazole anymore.
2 complexes of BATCl8 and the two cucurbiturils. Again, the triazole C–H signal is shifted downfield indicating that C–H⋯O hydrogen bonds are also formed in this complex. In contrast to BATCl8@CB74, however, no positionally shifted isomers are observed anymore. We conclude the CB8 host molecules to bind to the triazole C–H with one of their carbonyl oxygen atoms. Therefore, the CB7 molecules now prefer to be located centrally on the inner viologens and do not shift towards the triazole anymore.
Finally, the formation of the viologen–naphthalene charge-transfer complexes inside the CB8 cavities is also confirmed by the presence of the two typical absorption bands at 383 nm and 488 nm in the UV/Vis spectrum of the complex (Fig. 3e).19 These two bands are absent in BATCl8@CB74 and only appear in the presence of CB8.
All these results provide a picture not only consistent with the formation of a folded BATCl8@CB72·CB82 complex with CB8 in the peripheral and the CB7 in the central positions along the guest chain. It also nicely agrees with the details observed in the first step of the transformation cascade. The differences between both complexes in the tandem MS fragmentation and NMR experiments provide quite detailed insight into the structures of both complexes. It should be noted that the second step does not only comprise reorganization of the host–guest complex and refolding of the chain into a more contracted structure, but also the liberation of two empty CB7 molecules reminiscent of the formation of secondary messengers in signaling cascades in nature.
Third step: refolding through viologen cation-radical dimerization inside CB8
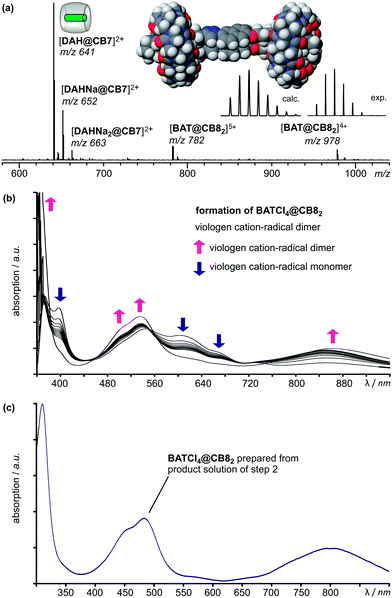
Initially, the third step was intended to lead to yet another foldamer of the guest chain using a redox instead of a chemical signal. It is well known that viologen dications can be reduced to the corresponding cation-radicals, which tend to dimerize in particular, when CB8 is present providing stabilization to the dimer by encapsulation. However, the first attempts to achieve a four-electron reduction of the guest chain with Zn powder failed and only incomplete reduction was obtained (see ESI†). Therefore, Na2S2O4 has been used as the reductant. But still, the presence of free CB7 in the sample solution caused incomplete refolding of the chain as CB7 binds to the viologen cation-radical monomer and thus competes with CB8 efficiently (see ESI†).28 Finally, the addition of the reductant together with 4 eq. 1,6-diaminohexane dihydrochloride DAHCl2 as a scavenger for all CB7 resulted in a clean transformation. The third step thus requires the simultaneous action of a redox and a chemical signal.When the sample solution obtained in step 2 is treated with 20 eq. of Na2S2O4 as the reductant and 4 eq. of DAHCl2, the ESI mass spectrum in Fig. 4a is obtained after a reaction time of 24 hours. Clearly, signals appear for the [DAH@CB7]2+ dication (m/z 641) accompanied by two signals in which one or two protons have been exchanged against sodium ions (m/z 652 and 663). The second species detected is indeed the quadruply reduced [BAT@CB82]4+ complex (m/z 978). As discussed above, one-electron oxidation reactions can occur at the ESI needle in the ion source. This would be one potential reason, why the [BAT@CB82]5+ ion (m/z 782) is also observed. A second possibility is of course the incomplete reduction of the viologens.
To distinguish both possibilities, the reduction was investigated by UV/Vis spectroscopy. First, an independent sample containing BATCl8 and 2 eq. of CB8 was prepared in water. Directly after the addition of Na2S2O4, the UV/Vis spectrum shows the presence of both the viologen cation-radical dimer with its typical bands at 367, 509, 538, and 865 nm and the viologen cation-radical monomer as indicated by the additional bands at 394 and 601 nm.14 Over time, the bands of the monomer decrease, while the ones of the dimer increase to reach complete conversion after ca. 2 hours. Usually, the dimerization of the cation-radical is a very fast process proceeding in a millisecond time regime.19 A control experiment with free BATCl8 confirmed this to be true for this guest, too. Immediately after the addition of the reductant, only cation-radical dimers were observed in the UV/Vis spectra (see ESI†). We therefore interpret our results for the BATCl8@CB82 complex as follows: before the reduction of the guest chain, a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex of CB8 and BATCl8 forms. As only the terminal viologens can form CT complexes with the adjacent naphthalenes, the CB8 molecules will certainly be located at the termini. Directly after the addition of reductant, the two inner viologens are quickly reduced and dimerize giving rise to the dimer bands in the UV/Vis spectra already right at the beginning of the experiment. The terminal viologens are, however, protected against reduction by CT complex formation and its stabilization by CB8 binding. This reduction proceeds thus much more slowly and stepwise so that the cation-radical monomer is observed as an intermediate. In the end, the monomers recombine into the folded structure shown in Fig. 4a, which contains two cation-radical dimers, both of which bind one CB8.
1 complex of CB8 and BATCl8 forms. As only the terminal viologens can form CT complexes with the adjacent naphthalenes, the CB8 molecules will certainly be located at the termini. Directly after the addition of reductant, the two inner viologens are quickly reduced and dimerize giving rise to the dimer bands in the UV/Vis spectra already right at the beginning of the experiment. The terminal viologens are, however, protected against reduction by CT complex formation and its stabilization by CB8 binding. This reduction proceeds thus much more slowly and stepwise so that the cation-radical monomer is observed as an intermediate. In the end, the monomers recombine into the folded structure shown in Fig. 4a, which contains two cation-radical dimers, both of which bind one CB8.
The UV/Vis spectrum of BATCl4@CB82 prepared by fully reducing a sample solution obtained from the second step of the cascade (figure c) clearly reveals the exclusive presence of cation-radical dimers, while the typical bands of the monomers are absent. Consequently, we conclude not only the [BAT@CB82]5+ ion in the ESI mass spectra to be generated during the electrospray process, but the third transformation step to proceed cleanly. The cooperation of a redox and a chemical signal give rise to yet another foldamer of the guest chain.
Forth step: going a step back by reoxidation and the addition of cucurbit[6]uril
Reoxidation of the guest chain is possible by bubbling air through the sample solution over a period of 10 minutes and the UV/Vis spectrum indicates that the cation-radical dimers are back-converted into viologen dications. Also, the bands typical for the charge-transfer complexes of the terminal viologens and the naphthalenes reappear (see ESI†). The simultaneous addition of 2 eq. of cucurbit[6]uril scavenges half of the DAHCl2 guests, which bind more strongly to CB6 than to CB7. Consequently, 2 eq. of CB7 become available again to reform the BATCl8@CB72·CB82 complex.The ESI mass spectrum in Fig. 5a clearly exhibits signals for all expected complexes and provides evidence for the simultaneous formation of DAHCl2@CB6 and DAHCl2@CB7 together with BATCl8@CB72·CB82, while other potentially possible complexes are not observed. The exclusive formation of these three complexes is also supported by the 1H,1H COSY NMR spectra (see ESI†), which very clearly show the formation of the viologen–naphthalene charge transfer complexes incorporated in BATCl8@CB72·CB82 as well as the formation of both DAHCl2@CB6 and DAHCl2@CB7 in equal amounts. The chemical shifts observed for the latter two complexes are in agreement with those reported earlier.13a
Fifth step: a complicated cucurbituril “metathesis”
Finally, the aim of the last step in the cascade reported here aims at the exchange of the two CB7 molecules located at the central viologens against two slightly weaker binding CB8 molecules to produce the BATCl8@CB84 complex. This goal can be achieved, when 2 eq. of CB8 are added simultaneously with 4 eq. ADACl as a preferred guest for CB7. As this will liberate two DAHCl2 guest dications from the remaining DAHCl2@CB7 complexes, it is furthermore required to add 2 eq. of CB6 in order to obtain a clean transformation.Fig. 5b shows the corresponding ESI mass spectrum, which – in view of the many different compounds now present in the mixture – is surprisingly clean and nicely confirms the self-sorting to work as expected as long as stoichiometry is carefully controlled. In the mass spectrum, DAHCl2 is only observed in a complex with CB6 (as [DAH@CB6]2+ at m/z 557) and ADACl only binds to CB7 (as [ADA@CB7]+ at m/z 1314). Besides these two ions, the only more intense ion corresponds to [BAT@CB84]8+. Again, 1H,1H COSY NMR spectra support these mass spectrometric results (see ESI†). While the signals for DAHCl2@CB6 prevail, those observed in step 4 for DAHCl2@CB7 vanish and new signals for ADACl@CB7 appear.13a Also, the typical chemical shifts of the viologens and naphthalenes forming the charge transfer complex are present. In conclusion, also the last transformation step in the cascade has successfully been carried out.
Conclusions
A transformation cascade has been presented which is reminiscent of natural signaling cascades although it only uses synthetic molecules. Nevertheless, conceptually, there are similarities: (i) a sequence-programmed guest chain is used instead of a biopolymer with sequence information. (ii) External signals result in molecular recognition events that at least in some of the steps of our cascade lead to conformational changes in the guest chain. Different foldamers are generated depending on the external stimuli. (iii) In some steps, secondary messengers are liberated that bind other molecules present in the mixture. As long as the stoichiometry is balanced, the self-sorting works nicely. (iv) Quite complex mixtures are generated, in which the guest selectivities of the three different cucurbiturils synergize with the sequence programmed into the guest chain and its rigidity-mediated folding properties to give rise to a surprisingly clear-cut behavior.Certainly, there are also significant differences between natural signalling cascades and that presented here. (i) While in our case, the system always finds its way back into thermodynamic equilibrium after addition of each of the chemical stimuli, natural cascades are kept far away from thermodynamic control by an energy flow through the living organism. (ii) Natural systems contain feedback loops. Together with the energy flow, which drives them, emergent properties arise that are much more complex than those observed here. (iii) Although our system is much less complex than natural systems, it is likely more sensitive to deviations from ideal stoichiometries.
Our study is a proof-of-principle study which paves the way into much more complex systems in the future. Many other steps can be imagined that could be performed with the system presented here. The diversity increases even more, when one considers guest chains with different and maybe longer sequences. This clearly indicates the potential of our approach for future work.
Acknowledgements
We thank Dr Andreas Springer and Fabian Klautzsch for help with the mass spectrometric experiments and Dr Andreas Schäfer and the staff of the BioSupraMol Core Facility for recording NMR spectra. We are grateful for funding from the Deutsche Forschungsgemeinschaft (SFB 765).Notes and references
- (a) R. A. R. Hunt and S. Otto, Chem. Commun., 2011, 47, 847–858 RSC; (b) G. von Kiedrowski, S. Otto and P. Herdewijn, J. Syst. Chem., 2010, 1, 1–6 CrossRef; (c) J. J. P. Peyralans and S. Otto, Curr. Opin. Chem. Biol., 2009, 13, 705–713 CrossRef CAS PubMed; (d) R. F. Ludlow and S. Otto, Chem. Soc. Rev., 2008, 37, 101–108 RSC.
- For selected examples of self-accelerating processes in synthetic chemical networks, see: (a) G. Ashkenasy, Z. Dadon, S. Alesebi, N. Wagner and N. Ashkenasy, Isr. J. Chem., 2011, 51, 106–117 CrossRef CAS; (b) A. Dieckmann, S. Beniken, C. D. Lorenz, N. L. Doltsinis and G. von Kiedrowski, Chem.–Eur. J., 2011, 17, 468–480 CrossRef CAS PubMed; (c) E. Kassianidis, R. J. Pearson, E. A. Wood and D. Philp, Faraday Discuss., 2010, 145, 235–254 RSC; (d) N. Wagner and G. Ashkenasy, Chem.–Eur. J., 2009, 15, 1765–1775 CrossRef CAS PubMed; (e) S. Kamioka, D. Ajami and J. Rebek Jr, Chem. Commun., 2009, 7324–7326 RSC; (f) D. G. Blackmond, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 5732–5736 CrossRef CAS PubMed; (g) K. Soai, T. Shibata and I. Sato, Acc. Chem. Res., 2000, 33, 382–390 CrossRef CAS PubMed; (h) D. H. Lee, K. Severin, Y. Yokobayashi and M. R. Ghadiri, Nature, 1997, 390, 591–594 CrossRef CAS PubMed; (i) K. Severin, D. H. Lee, J. A. Martinez, M. Vieth and M. R. Ghadiri, Angew. Chem., Int. Ed., 1998, 37, 126–128 CrossRef CAS.
- D. Newth and J. Finnigan, Aust. J. Chem., 2006, 59, 841–848 CrossRef CAS.
- (a) A. J. Pross, J. Syst. Chem., 2011, 2, 1–14 CrossRef CAS; (b) J. W. Szostak, Nature, 2009, 459, 171–172 CrossRef CAS PubMed; (c) A. Pross, Chem.–Eur. J., 2009, 15, 8374–8381 CrossRef CAS PubMed; (d) S. Kauffman, At Home in the Universe, Oxford University Press, New York, 1996 Search PubMed; (e) S. Kauffman, Origins of Order: Self-Organization and Selection in Evolution, Oxford University Press, Oxford, 1993 Search PubMed.
- (a) S. Rasmussen, M. Bedau, L. Chen, D. Deamer, D. Krakauer, N. Packard and P. Stadler, Protocells: Bridging Nonliving and Living Matter, MIT Press, Cambridge, 2009 Search PubMed; (b) P. L. Luisi, The Emergence of Life: From Chemical Origins to Synthetic Biology, Cambridge University Press, Cambridge, 2006 Search PubMed; (c) J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS PubMed.
- (a) M. M. Safont-Sempere, G. Fernández and F. Würthner, Chem. Rev., 2011, 111, 5784–5814 CrossRef CAS PubMed; (b) C. Talotta, C. Gaeta, Z. Qi, C. A. Schalley and P. Neri, Angew. Chem., Int. Ed., 2013, 52, 7437–7441 CrossRef CAS PubMed; (c) K. Mahata, M. L. Saha and M. Schmittel, J. Am. Chem. Soc., 2010, 132, 15933–15935 CrossRef CAS PubMed; (d) W. Jiang, A. Schäfer, P. C. Mohr and C. A. Schalley, J. Am. Chem. Soc., 2010, 132, 2309–2320 CrossRef CAS PubMed; (e) K. Mahata and M. Schmittel, J. Am. Chem. Soc., 2009, 131, 16544–16554 CrossRef CAS PubMed; (f) W. Jiang and C. A. Schalley, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10425–10429 CrossRef CAS PubMed; (g) Y. Rudzevich, V. Rudzevich, F. Klautzsch, C. A. Schalley and V. Böhmer, Angew. Chem., Int. Ed., 2009, 48, 3867–3871 CrossRef CAS PubMed; (h) P. Mukhopadhyay, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2006, 128, 14093–14102 CrossRef CAS PubMed.
- (a) I. Saur, R. Scopelliti and K. Severin, Chem.–Eur. J., 2006, 12, 1058–1066 CrossRef CAS PubMed; (b) K. Severin, Chem.–Eur. J., 2004, 10, 2565–2580 CrossRef CAS PubMed; (c) Z. Grote, R. Scopelliti and K. Severin, Angew. Chem., Int. Ed., 2003, 42, 3821–3825 CrossRef PubMed.
- V. Del Amo and D. Philp, Chem.–Eur. J., 2010, 16, 13304–13318 CrossRef CAS PubMed.
- J. M. A. Carnall, C. A. Waudby, A. M. Belenguer, M. C. A. Stuart, J. J. P. Peyralans and S. Otto, Science, 2010, 327, 1502–1506 CrossRef CAS PubMed.
- (a) V. E. Campbell, X. de Hatten, N. Delsuc, B. Kauffmann, I. Huc and J. R. Nitschke, Nat. Chem., 2010, 2, 684–687 CrossRef CAS PubMed; (b) V. E. Campbell, X. de Hatten, N. Delsuc, B. Kauffmann, I. Huc and J. R. Nitschke, Chem.–Eur. J., 2009, 15, 6138–6142 CrossRef CAS PubMed.
- (a) E. Masson, X. Ling, J. Roymon, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC; (b) K. Kim, Chem. Soc. Rev., 2002, 31, 96–107 RSC.
- (a) E. Masson, X. Lu, X. Ling and D. L. Patchell, Org. Lett., 2009, 11, 3798–3801 CrossRef CAS PubMed; (b) M. V. Rekharsky, T. Mori, C. Yang, Y. H. Ko, N. Selvapalam, H. Kim, D. Sobransingh, A. E. Kaifer, S. Liu, L. Isaacs, W. Chen, S. Moghaddam, M. K. Gilson, K. Kim and Y. Inoue, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20737–20742 CrossRef CAS PubMed; (c) W. S. Jeon, K. Moon, S. H. Park, H. Chun, Y. H. Ko, J. Y. Lee, E. S. Lee, S. Samal, N. Selvapalam, M. V. Rekharsky, V. Sindelar, D. Sobransingh, Y. Inoue, A. E. Kaifer and K. Kim, J. Am. Chem. Soc., 2005, 127, 12984–12989 CrossRef CAS PubMed; (d) J. Mohanty and W. M. Nau, Angew. Chem., Int. Ed., 2005, 44, 3750–3754 CrossRef CAS PubMed.
- (a) S. Liu, C. Ruspic, P. Mukhopadhyay, S. Chakrabarti, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2005, 127, 15959–15967 CrossRef CAS PubMed; (b) K. Kim, N. Selvapalam and D. H. Oh, J. Inclusion Phenom. Macrocyclic Chem., 2004, 50, 31–36 CrossRef CAS.
- (a) Z. Zhang, Y. Zhang and L. Yu, J. Org. Chem., 2011, 76, 4682–4685 CrossRef CAS PubMed; (b) H. Zhang, Q. Wang, M. Liu, X. Ma and T. He, Org. Lett., 2009, 11, 3234–3237 CrossRef CAS PubMed; (c) W. M. Nau, G. Ghale, A. Hennig, H. Bakirci and D. M. Bailey, J. Am. Chem. Soc., 2009, 131, 11558–11570 CrossRef CAS PubMed.
- (a) G. Celtek, M. Artar, O. A. Scherman and D. Tuncel, Chem.–Eur. J., 2009, 15, 10360–10363 CrossRef CAS PubMed; (b) S.-Y. Kim, Y. H. Ko, J. W. Lee, S. Sakamoto, K. Yamaguchi and K. Kim, Chem. – Asian J., 2007, 2, 747–754 CrossRef CAS PubMed; (c) P. Mukhopadhyay, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2006, 128, 14093–14102 CrossRef CAS PubMed; (d) P. Mukhopadhyay, A. Wu and L. Isaacs, J. Org. Chem., 2004, 69, 6157–6164 CrossRef CAS PubMed.
- W. Jiang, W. Qi, I. Linder, F. Klautzsch and C. A. Schalley, Chem.–Eur. J., 2011, 17, 2344–2348 CrossRef CAS PubMed.
- F. Biedermann and O. A. Scherman, J. Phys. Chem. B, 2012, 116, 2842–2849 CrossRef CAS PubMed.
- W. S. Jeon, A. Y. Ziganshina, J. W. Lee, Y. H. Ko, J.-K. Kang, C. Lee and K. Kim, Angew. Chem., Int. Ed., 2003, 42, 4097–4100 CrossRef CAS PubMed.
- (a) W. S. Jeon, E. Kim, Y. H. Ko, I. Hwang, J. W. Lee, S.-Y. Kim, H.-J. Kim and K. Kim, Angew. Chem., Int. Ed., 2005, 44, 87–91 CrossRef CAS PubMed.
- P. Neta and M. C. Richoux, J. Chem. Soc., Faraday Trans. 2, 1985, 81, 1427–1443 RSC.
- C. A. Schalley, C. Verhaelen, F.-G. Klärner, U. Hahn and F. Vögtle, Angew. Chem., Int. Ed., 2005, 44, 480–484 CrossRef PubMed.
- H.-J. Kim, W. S. Jeon, Y. H. Ko and K. Kim, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5007–5011 CrossRef CAS PubMed.
- There is precedent for the loss of N2 from triazole rings: G. Maier, J. Eckwert, A. Bothur, H. P. Reisenauer and C. Schmidt, Liebigs Ann., 1996, 1041–1053 CAS.
- For a review on C–H⋯O hydrogen bonds, see: (a) R. Castellano, Curr. Org. Chem., 2004, 8, 845–865 CrossRef CAS . Two recent examples for C–H⋯X hydrogen bonds involving triazoles; (b) E. V. Dzyuba, L. Kaufmann, N. L. Löw, A. K. Meyer, H. D. F. Winkler, K. Rissanen and C. A. Schalley, Org. Lett., 2011, 13, 4838–4841 CrossRef CAS PubMed; (c) S. C. Picot, B. R. Mullaney and P. D. Beer, Chem.–Eur. J., 2012, 18, 6230–6237 CrossRef CAS PubMed.
- M. K. Sinha, O. Reany, M. Yefet, M. Botoshanskyand and E. Keinan, Chem.–Eur. J., 2012, 18, 5589–5605 CrossRef CAS PubMed.
- W. S. Jeon, H.-J. Kim, C. Lee and K. Kim, Chem. Commun., 2002, 1828–1829 RSC.
- Y. H. Ko, E. Kim, I. Hwang and K. Kim, Chem. Commun., 2007, 1305–1315 RSC.
- (a) W. J. Lee, S. Samal, N. Selvapalam, H.-J. Kim and K. Kim, Acc. Chem. Res., 2003, 36, 621–630 CrossRef PubMed; (b) W. S. Jeon, H.-J. Kim, C. Lee and K. Kim, Chem. Commun., 2002, 1828–1829 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Complete experimental, synthetic procedures and analytical data of new compounds, additional NMR and MS experiments and control experiments. See DOI: 10.1039/c3sc53211a |
| This journal is © The Royal Society of Chemistry 2014 |