Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster†
John D.
Sidda
ab,
Lijiang
Song
a,
Vincent
Poon
ab,
Mahmoud
Al-Bassam
c,
Orestis
Lazos
a,
Mark J.
Buttner
c,
Gregory L.
Challis
a and
Christophe
Corre
*ab
aDepartment of Chemistry, University of Warwick, Coventry, CV4 7AL, UK. E-mail: C.Corre@warwick.ac.uk; Fax: +44 (0)2476 524 112; Tel: +44 (0)2476 523 557
bSchool of Life Sciences, University of Warwick, Coventry, CV4 7AL, UK
cDepartment of Molecular Microbiology, John Innes Centre, Norwich, NR4 7UH, UK
First published on 30th October 2013
Abstract
Gaburedins, a family of γ-aminobutyrate (GABA)-derived ureas, have been discovered by deletion of gbnR, an arpA-like putative transcriptional repressor in Streptomyces venezuelae ATCC 10712. Comparison of metabolite profiles in the wild type and mutant strains revealed six metabolites in the mutant that are lacking from the wild type. The structure of gaburedin A was established by HRMS combined with 1- and 2-D NMR spectroscopy and was confirmed by total synthesis. The other metabolites were confirmed as congeners using HRMS, MS/MS and feeding of putative biosynthetic precursors. Two genes, gbnA and gbnB, are proposed to be involved in gaburedin biosynthesis. Consistent with this hypothesis, deletion of gbnB in the gbnR mutant abolished gaburedin production. This is the first report to disclose the discovery of novel natural products via rational deletion of a putative pathway-specific regulatory gene.
Streptomyces are soil-dwelling, filamentous bacteria that produce a remarkably diverse range of bioactive natural products. Such molecules find use in the treatment of infectious diseases, cancer and transplant rejection, as well as in agriculture. Indeed, almost 70% of commercially-available antibiotics originate from the Streptomyces genus.1 Classical methods for identification of novel natural products have focused on screening cultures for specific bioactivities. However, these approaches often result in the re-discovery of known compounds. Alternative strategies are therefore required for the discovery of novel bioactive natural products from microbial sources.2
The recent explosion in the availability of bacterial genome sequences has led to the identification of many cryptic natural product biosynthetic gene clusters.3 These offer a promising resource for novel natural product discovery.3 However, many cryptic biosynthetic gene clusters are expressed poorly or not at all under laboratory growth conditions.4,5 Rational approaches are therefore required to induce the expression of such silent biosynthetic pathways.
In addition to biosynthetic and self-resistance genes, single or multiple genes encoding transcriptional regulators (activators or repressors) are typically present in bacterial natural product biosynthetic gene clusters.5 Recently, it has been reported that the constitutive expression of a putative pathway-specific transcriptional activator results in the production of a novel complex of macrolide antibiotics by S. ambofaciens.4 A similar strategy has been employed in Aspergillus nidulans to discover the product of a cryptic biosynthetic pathway.6 Deletion of putative bacterial pathway-specific transcriptional repressors has been investigated as a complementary approach, but has yet to result in the discovery of novel metabolites.7,8 ArpA-like transcriptional repressors belong to the TetR-family of DNA-binding proteins and are known to control the production of a wide variety of antibiotics.9,10 ArpA itself controls the production of streptomycin in Streptomyces griseus.11 The biosynthesis of several other natural products is repressed by ArpA-like proteins and inactivation of arpA-like genes generally boosts production levels, e.g. deletion of mmyR in Streptomyces coelicolor has been shown to result in the overproduction of methylenomycin antibiotics.12
ArpA-like proteins have been shown to sense and respond to specific ligands such as γ-butyrolactones, 2-alkyl-4-hydroxymethylfuran-3-carboxylic acids (AHFCAs) and butenolides.13–17 In particular, the methylenomycin furans, assembled by the products of the mmfLHP genes, act as methylenomycin production inducers in S. coelicolor. These signaling molecules are thought to interact with the ArpA-like transcriptional repressors MmfR and/or MmyR.12,14 Even though MmfR and MmyR share 35% identity and 56% similarity, the phenotypes of mmfR and mmyR mutants are distinct. The strain lacking mmyR overproduces the methylenomycin antibiotics, whereas the strain lacking mmfR exhibits a phenotype similar to the wild type strain.12
BLAST searches revealed that several Streptomyces genomes contain a cassette of five genes homologous to mmfR–mmfLHP–mmyR. These are proposed to be involved in the transcriptional regulation of specific natural product biosynthetic pathways.14 Thus, inactivation of the mmyR homologues in these genomes was predicted to result in induction of the biosynthetic gene clusters under their control and biosynthesis of the corresponding metabolic products.
The genome sequence of Streptomyces venezuelae ATCC 10712 became publicly available in 2012, revealing a cluster of genes that contains the mmyR homologue gbnR (sven_4187), as well as the mmfR homologue sgnR (sven_4182) and the mmfLHP homologues sgnLPH (sven_4183, 4185 and 4188; Fig. 1). This putative regulatory system is located adjacent to gbnABC (sven_4179–4181), which encode potential natural product biosynthetic and export proteins.
 | ||
| Fig. 1 Organisation of the gbn/sgn gene cluster (9.2 kb) that regulates and directs gaburedin production in S. venezuelae ATCC 10712 and proposed functions of the encoded proteins. | ||
In silico analysis of the gbn gene cluster revealed highly conserved DNA sequences in the sgnR-sgnL and gbnR-sgnH intergenic regions as well as in the promoter region of gbnA (Fig. 1). These conserved DNA sequences are proposed to be binding sites for GbnR (Fig. 1). As a consequence, GbnR was predicted to act as a repressor of the gbnABC operon, which should therefore be constitutively expressed if gbnR is deleted.
Here we report that inactivation of gbnR in S. venezuelae induces production of the gaburedins, a novel family of γ-aminobutyrate ureas. To the best of our knowledge, this is the first example of the discovery of a novel family of bacterial natural products via rational manipulation of a putative pathway-specific repressor gene.
The S. venezuelae gbnR::apr mutant was constructed using PCR-targeting (see ESI†).18 The gbnR::apr and wild type strains were grown on a supplemented minimal agar medium for 3 days, after which the acidified agar medium was extracted with ethyl acetate. The extracts were dried and re-suspended in water–methanol (1:1). Metabolites produced by the wild type and mutant strains were analysed by high resolution LC-MS/MS.
Comparison of the base peak chromatograms revealed six metabolites (1–6), named gaburedins, which were produced by the gbnR::apr mutant strain but not the wild type strain (Fig. 2).
The molecular formulae of the metabolites were deduced to be C14H18N2O5 for 1, C11H20N2O5 for 2 and 3, C10H18N2O5 for 4, C10H16N2O6S for 5 and C10H18N2O5S for 6. A neutral loss of 129 Da was observed for each parent ion in the MS/MS spectrum, corresponding to loss of a common C5H7NO3 fragment (Fig. 3).
 | ||
| Fig. 3 High resolution MS data for gaburedin D (4), C (3), B (2) and A (1) (top panel to bottom panel, respectively), highlighting the neutral loss of 129 Da observed for all gaburedins. | ||
Interestingly, the molecular formulae of the resulting daughter ions matched those of the protonated amino acids phenylalanine, isoleucine/leucine, valine and methionine for compounds 1, 2/3, 4 (Fig. 5) and 6 respectively. For compound 5, the molecular formulae for the daughter ion matched that of acetylcysteine (C5H10NO3S). A further loss of a 42 Da fragment from this daughter ion was consistent with the presence of an acetyl group. An analogous fragmentation pattern was observed for the [M + Na]+ ions of each gaburedin.
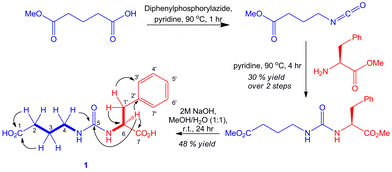
Cultures of the gbnR::apr mutant were scaled up to isolate sufficient quantities of 1 for structure elucidation by NMR spectroscopy (Fig. 4). COSY and HMBC experiments allowed unambiguous elucidation of the planar structure. An authentic standard of S-1 was synthesised via modification of the route reported by Tayaka and co-workers for the synthesis of ureido-containing derivatives of 5-fluorouracil (Fig. 4).19 LC-MS/MS and NMR analyses confirmed that the compound purified from the gbnR::apr mutant strain was identical to the synthetic standard 1.
 | ||
| Fig. 5 Structures of gaburedins A–D (1–4) isolated from the S. venezuelae gbnR::apr mutant, showing the conserved GABA fragment (boxed) common to all gaburedins. | ||
Supplementation of the culture medium with specific amino acids was found to influence gaburedin production. For example, growth of the gbnR::apr mutant in a culture medium enriched in L-phenylalanine (20 mM final concentration) resulted in overproduction of gaburedin A 1 relative to the other gaburedin congeners. Feeding with D-phenylalanine resulted in the overproduction of a gaburedin with same retention time and fragmentation pattern as 1. However, chiral HPLC analysis indicated that this compound was the enantiomer of 1, confirming that the absolute configuration of 1 initially isolated from S. venezuelae is S (ESI†).
The gbnR::apr mutant was grown in culture media supplemented with either L-isoleucine or L-leucine (20 mM final concentration) to distinguish between gaburedin B 2 and gaburedin C 3 which have very similar retention times (Fig. 2). The resulting culture extracts were analysed by LC-MS/MS, revealing that the quantity of 3 increased dramatically when L-isoleucine was fed, whereas the quantity of 2 significantly increased in the culture extract obtained from the mutant grown in the presence of L-leucine (see ESI†). Specific incorporation of d10-leucine into 2 and d10-isoleucine into 3 further confirmed the structural identity of these metabolites (ESI†). Similarly the gbnR::apr mutant strain was grown in the presence of N-acetylcysteine, in which case gaburedin E 5 was overproduced. Further precursor-directed biosynthesis experiments were carried out by supplementing the culture medium with various amino acids and gaburedins containing Val, Met, Tyr, Trp, Gly, Ala, Pro, Ser, Thr and Glu were identified by LC-MS analyses (see ESI†). The molecular formulae, fragmentation patterns and retention times were all consistent with the proposed structures (see ESI†). Interestingly supplementation with γ-aminobutyrate (GABA) resulted in the production of a GABA dimer (7), in which the two monomers are linked by a ureido bridge (molecular formulae C9H16N2O5, Scheme 1). Amino acid analogues lacking the carboxylic acid group were also incorporated into gaburedin analogues, e.g. isobutylamine was found to elicit the production of compound 8 (Scheme 1).
The gbnA gene encodes a putative pyridoxal phosphate (PLP)-dependent amino acid decarboxylase (Fig. 1), which is proposed to decarboxylate glutamate to give GABA (Scheme 1). Interestingly, feeding of the S. venezuelae gbnR mutant with glutamate not only yielded the glutamate-derived gaburedin analogue but also analogue 7, consistent with the hypothesis that GABA is derived from decarboxylation of glutamate. The gbnB gene encodes a putative ATP-dependent enzyme belonging to the acyl-CoA synthetase superfamily (Fig. 1). GbnB is proposed to adenylate either the carboxyl or the carbamoyl group of compound 9, which derives from the spontaneous reaction of GABA with CO2 (Scheme 1). Carboxyl adenylate 10 would need to undergo cyclisation to the N-carboxyanhydride 11, which would then afford the gaburedins via ring opening with amino acids (note, however, that this would involve nucleophilic attack at the less electrophilic carbonyl group of 11, which is chemically unlikely). On the other hand, the carbamoyl adenylate 12 could react with the appropriate amino acids either directly, or via anhydride 11, to form the gaburedins. Deletion of gbnB in the gbnR mutant of S. venezuelae abolished gaburedin production (Fig. 2), consistent with the key role proposed for GbnB in gaburedin biosynthesis. GbnA and GbnB appear to be the only two enzymes required for gaburedin biosynthesis. GbnC is proposed to export either the gaburedins or intermediate 11/12, which could react with amino acids in the extracellular milieu to form the gaburedins.
A handful of other urea-containing natural products are known, such as the syringolins and the pacidamycins.20–22 Interestingly, the adenylation domain of SylC, which has been shown to catalyze urea formation in syringolin biosynthesis, shares 39% similarity and 22% identity with GbnB.20 The mechanism for SylC-catalyzed urea formation has been proposed to involve a five-membered cyclic N-carboxyanhydride, or a closely related species.20 This anhydride is hypothesized to be formed via an analogous mechanism to that suggested for the formation of 11 (Scheme 1). Trapping of the anhydride with an isoleucyl or valinyl acyl carrier protein thioester would lead to formation of the ureido linkage of the syringolins. As noted above, an analogous mechanism for the formation of the gaburedins via intermediate 11 seems unlikely. Thus, further experiments are required to establish whether similar or distinct mechanisms are utilized for urea formation in syringolin and gaburedin biosynthesis.
Interestingly, BLAST searches revealed analogous operons to the gbnABC operon in other bacteria such as Streptococcus mutans, Salmonella enterica and Vibrio vulnificus (see ESI†). Surprisingly, most of these are clinical isolates of opportunistic pathogens. It will be important, therefore, to establish the nature of the metabolites produced by these strains and to investigate the role of gaburedins and related molecules in pathogenicity.
Conclusions
In summary, bioinformatics analyses suggested that expression of the gbnABC gene is controlled by the putative pathway-specific transcriptional repressor GbnR in S. venezuelae. Inactivation of gbnR resulted in derepression of the gbn gene cluster, resulting in the discovery of the gaburedins, a novel class of urea natural products. To the best of our knowledge, this is the first report of novel natural product discovery via rational manipulation of a pathway specific transcriptional repressor gene, an approach that offers considerable potential for activation of silent biosynthetic pathways in Streptomyces species, which could lead to the discovery of a wide variety of new metabolites.Acknowledgements
This work was mainly supported by a University Research Fellowship (UF090255) from The Royal Society and by an EPSRC DTA studentship to J. D. Sidda. The Bruker MaXis mass spectrometer used in this research was obtained with support from AWM and the ERDF. The authors would also like to thank Dr David J. Fox for helpful discussions.Notes and references
- D. A. Hopwood, Streptomyces in Nature and Medicine: The Antibiotic Makers, Oxford University Press, New York, 2007 Search PubMed.
- F. E. Koehn and G. T. Carter, Nat. Rev. Drug Discovery, 2005, 4, 206–220 CrossRef CAS PubMed.
- M. Zerikly and G. L. Challis, ChemBioChem, 2009, 10, 625–633 CrossRef CAS PubMed.
- L. Laureti, L. Song, S. Huang, C. Corre, P. Leblond, G. L. Challis and B. Aigle, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6258–6263 CrossRef CAS PubMed.
- G. Liu, K. F. Chater, G. Chandra, G. Niu and H. Tan, Microbiol. Mol. Biol. Rev., 2013, 77, 112–143 CrossRef CAS PubMed.
- S. Bergmann, J. Schümann, K. Scherlach, C. Lange, A. A. Brakhage and C. Hertweck, Nat. Chem. Biol., 2007, 3, 213–217 CrossRef CAS PubMed.
- M. Gottelt, S. Kol, J. P. Gomez-Escribano, M. J. Bibb and E. Takano, Microbiology, 2010, 156, 2243–2253 CrossRef PubMed.
- R. Bunet, L. Song, M. V. Mendes, C. Corre, L. Hotel, N. Rouhier, X. Framboisier, P. Leblond, G. L. Challis and B. Aigle, J. Bacteriol., 2011, 193, 1142–1153 CrossRef CAS PubMed.
- J. L. Ramos, M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan and R. Tobes, Microbiol. Mol. Biol. Rev., 2005, 69, 326–356 CrossRef CAS PubMed.
- L. Cuthbertson and J. R. Nodwell, Microbiol. Mol. Biol. Rev., 2013, 77, 440–475 CrossRef CAS PubMed.
- Y. Ohnishi, S. Kameyama, H. Onaka and S. Horinouchi, Mol. Microbiol., 1999, 34, 102–111 CrossRef CAS.
- S. O'Rourke, A. Wietzorrek, K. Fowler, C. Corre, G. L. Challis and K. F. Chater, Mol. Microbiol., 2009, 71, 763–778 CrossRef CAS PubMed.
- H. Nishida, Y. Ohnishi, S. Horinouchi and T. Beppu, Environ. Microbiol., 2007, 9, 1986–1994 CrossRef CAS PubMed.
- C. Corre, L. Song, S. O'Rourke, K. F. Chater and G. L. Challis, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17510–17515 CrossRef CAS PubMed.
- S. Kitani, K. T. Miyamoto, S. Takamatsu, E. Herawati, H. Iguchi, K. Nishitomi, M. Uchida, T. Nagamitsu, S. Omura, H. Ikeda and T. Nihira, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16410–16415 CrossRef CAS PubMed.
- J. M. Willey and A. A. Gaskell, Chem. Rev., 2011, 111, 174–187 CrossRef CAS PubMed.
- K. Arakawa, N. Tsuda, A. Taniguchi and H. Kinashi, ChemBioChem, 2012, 13, 1447–1457 CrossRef CAS PubMed.
- B. Gust, G. L. Challis, K. Fowler, T. Kieser and K. F. Chater, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1541–1546 CrossRef CAS PubMed.
- T. Tayaka and Z. Tozuka, US Pat., 4,349,552, 14 September 1982.
- H. J. Imker, C. T. Walsh and W. M. Wuest, J. Am. Chem. Soc., 2009, 131, 18263–18265 CrossRef CAS PubMed.
- E. J. Rackham, S. Grüschow, A. E. Ragab, S. Dickens and R. J. M. Goss, ChemBioChem, 2010, 11, 1700–1709 CrossRef CAS PubMed.
- W. Zhang, I. Ntai, M. L. Bolla, S. J. Malcolmson, D. Kahne, N. L. Kelleher and C. T. Walsh, J. Am. Chem. Soc., 2011, 133, 5240–5243 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Strains and plasmids, experimental procedures, analytical data. See DOI: 10.1039/c3sc52536h |
| This journal is © The Royal Society of Chemistry 2014 |