Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Expression of Arabidopsis thaliana S-ACP-DES3 in Escherichia coli for high-performance biodiesel production
Barbara
Scaglia
a,
Elena
Cassani
b,
Roberto
Pilu
*a and
Fabrizio
Adani
*a
aDi.S.A.A. – Gruppo Ricicla – Biomass and Bioenergy Laboratory – University of Milan, Via Celoria 2, 20133, Milan, Italy. E-mail: salvatore.pilu@unimi.it; fabrizio.adani@unimi.it; Fax: +39-2-50316521/+39-2-50316521; Tel: +39-2-50316549/+39-2-50316547
bDi.S.A.A. – Genetic Laboratory – University of Milan, Via Celoria 2, 20133, Milan, Italy
First published on 11th November 2014
Abstract
The chemical characteristics of oil feedstocks greatly affect the physical properties of biodiesel. Bacteria, because of their very high growth-rate and their easy culture, can be used for making oil-feedstocks to produce biodiesel. In this way genetic transformation was recently applied to microorganisms to improve oil quality and hence biodiesel quality. In this work, Escherichia coli was genetically transformed by heterologous expression of Arabidopsis thaliana stearoyl-acyl carrier protein desaturase3 (S-ACP-DES3): the cDNA coding for S-ACP-DES3 from Arabidopsis thaliana was cloned into the pET-15b vector and expressed in the E. coli BL21 (DE3) strain. The S-ACP-DES3 protein obtained was expressed in a soluble form after induction with IPTG and visualized by SDS-PAGE analysis. The recombinant Escherichia coli fatty acid profile showed an optimal unsaturated vs. monosaturated and saturated FAs combination. These results can be used as a starting point to try to modify oleaginous bacteria to get both high oil productivity and optimal oil composition.
1. Introduction
Biodiesel is a fuel composed of fatty acid methyl esters (FAME) resulting from trans-esterification of fats or oils originating from different renewable sources.1 The literature indicates that first- and second-generation biodiesels are produced, respectively, starting from either food-plants (cereal crops, oil crops and sugar crops) or non-food-plant raw materials.2–4The rapid growth in demand for biodiesel necessitates finding alternative oil sources: the extraction of oil/fat from waste and by-products (e.g. animal fat from slaughter industry, soap-stock from vegetable oil and milk transformation industries) allow us to obtain biodiesel, reducing land-use and plant (i.e. edible and non-edible oleaginous crops) cultivation costs.3,5–7 Moreover, microbial oleaginous microorganisms (i.e. microalgae, fungi, yeast and bacteria) have all been considered as feedstocks to produce biodiesel,8 because of their short life, low labor requirement, and the fact that they are less affected by venue, season and climate; in addition they are easy to scale up.8,9
Depending on the starting biomass, biodiesel does not show a constant composition of fatty acids and as consequence of that, biodiesel physical properties are not constant. In particular the chain-length of fatty acids, the ratio between unsaturated and saturated fatty acids and the degree of unsaturation (mono, and poly-unsaturated fatty acids), are all chemical characteristics that influence greatly the biodiesel's physical properties.
Pour point, cloud point, flash point, and kinematic viscosity, all improve with the increasing of the degree of unsaturation (e.g. oil from corn, cottonseed, soybean and sunflower).9–11
In addition biodiesel that contains high concentrations of saturated fatty acids (such as those derived from coconut, palm and tallow, animal fat) excel in both cetane number and oxidation stability.11,12
Chemical composition also affects biodiesel's environmental impacts:9 biodiesel GHG and NOx gas emissions are usually lower than those of diesel. On the other hand, with the increasing of the number of double bonds in alkyl chains there is an augmentation of NOx production during biodiesel combustion. Therefore it becomes very difficult to define what are the best characteristics of the oils from which biodiesel is produced. A possible useful compromise is that suggested by some authors10,12 who propose using an oil-feedstock composed of both a high concentration of monounsaturated fatty acid (e.g. palmitoleic and oleic acids) and low concentrations of both saturated and polyunsaturated fatty acids. Moser and Vaughn13 gave more precise indications on optimal fatty acid composition to produce biodiesel, suggesting maximum/minimum concentration of each class of fatty acid with respect to the total volatile fatty acid content (FA): (i) saturated fatty acid (SFA) < 26% FA, (ii) m-unsaturated fatty acid (m-SFA) > 62% FA, (iii) trienoic fatty acid (TFA) < 7% FA, and, (iv) C20 FA + very long chain fatty acid (C20 + VLCFA) < 19% FA.
Bacteria rarely accumulate high amounts of oil; nevertheless their very high growth-rate and their easy culture methods suggest their use as good potential oil-feedstocks to produce biodiesel. For example the bacteria Escherichia coli (E. coli), is one of the best characterized and manipulable microorganism. It is approximately composed of 9–10% (w/w) lipid, reaching at industrial scale, a fatty acids productivity of 0.2 g l−1 h−1 per gram of cell mass.9
Aiming to obtain optimal oil composition, genetic engineering was applied to microorganisms to obtain a higher quality of marketable biodiesel.14,15 In the past, researchers' activities were addressed towards increasing fatty acid production but, also, to obtain an optimal fatty acid composition (i.e. an increase in the unsaturated fatty acid content). Lu et al.14 obtained a significant overproduction of fatty acid (19 times more than the control strain) by performing simultaneously the following genetic alterations: (i) knocking out the endogenous fadD gene, (ii) heterologous expression of a Cinnamomum camphorum seed thioesterase (iii) over-expression of acetyl-CoA carboxylase; (iv) over-expression of the endogenous thioesterase. Nevertheless the oil obtained did not have optimal characteristics, being rich in saturated fatty acids. In a different team, Cao et al.15 by homologous over expression of fabA and fabB genes plus the introduction of Arabidopsis thaliana thioesterase, obtained both an overproduction of fatty acid (+37.5% with respect to the control strain) as well as an increase in the unsaturated fatty acids content (UFA). However the modified oil showed high SFA and low UFA concentrations, that were not in line with the optimal oil composition as indicated above.13
Fatty acid desaturation in E. coli is controlled by fatty acid desaturase proteins; these proteins can be divided into three classes: acyl-CoA, acyl-ACP and acyl-lipid desaturases that comprise acyl-[acyl carrier protein] (ACP) desaturases, soluble enzymes able to catalyse the insertion of a double bond into saturated fatty acid bound to ACP.15 For example the Δ9 desaturases or Δ9-stearoyl (18:0)-ACP desaturase (S-ACP-DES, EC 1.14.99.6) is a ubiquitous plastid enzyme that converts stearic acid to oleic acid (18:1).16,17 S-ACP-DES was identified in several plants; in particular the Arabidopsis thaliana genome encodes 7 highly conserved S-ACP-DES-like enzymes that show different tissue-specific expression.17 The isoform S-ACP-DES3 (EC 1.14.19.2; http://www.brenda-enzymes) shows high expression in leaves and preferentially desaturates palmitoyl-acyl-carrier protein (16:0-ACP) substrate at the C9 position to the product palmitoleoyl-acyl-carrier protein.15,16
This work represents a first approach in improving desaturation of fatty acids of E. coli BL21 (DE3) strain, by expressing the S-ACP-DES3 gene cloned from Arabidopsis thaliana. This approach therefore represents a model that can be applied to oleaginous microorganisms in order to improve the quality of the biodiesel eventually produced.
2. Methods and materials
2.1 Strains, plasmid construction and cloning
With the aim of isolating the DES3 sequence from Arabidopsis thaliana (At5g16230), total RNA was isolated from leaves using the method described by van Tunen et al.18 RNA extracted was treated with DNAse I 1 U μl−1 (Deoxyribonuclease I, Amplification Grade, Invitrogen) for eliminating DNA during RNA purification procedures. First strand cDNA was synthesized with an oligo(dT) primer from total RNA using the Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen). First-strand cDNA was used as the template for PCR amplification. Amplification reactions were carried out on samples containing an aliquot of cDNA synthesized from 5 μg of total RNA, 5× Green Reaction Buffer, 2.5 mM MgCl2, 200 μM each dATP, dCTP, dGTP, and dTTP, 0.1 μM each primer, and 1 unit of GoTaq DNA Polymerase/Pfu Polymerase and were performed in a final volume of 50 μl. For DES3 amplification (accession number NM_121628) DES3F (5′-![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCGATGGCTTTGCTTTTGACA-3′; Tm 60 °C; position + 4 respect to the start codon) and DES3R (5′-
TCGATGGCTTTGCTTTTGACA-3′; Tm 60 °C; position + 4 respect to the start codon) and DES3R (5′-![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CTATAGTTTCACATCTCTACC-3′; Tm 58 °C; position + 1206 respect to the start codon). Both primers were designed containing an adaptor with the BamHI restriction site. The length of the amplified product was 1203 bp. The recovery of the amplified product from agarose gel was obtained with Freeze 'N Squeeze DNA Gel Extraction Spin Columns made by Bio-Rad (Hercules, California). It was used pCR4-TOPO vector of 3956 bp (Invitrogen, Carlsbad, California) and Escherichia coli One Shot TOP10 Kit (Invitrogen, Carlsbad, California) to clone the DES3 sequence isolated as described above. The identity of the products was confirmed by sequencing. The isolation of plasmid pCR4-TOPO-DES3 DNA was obtained from QIAGEN Plasmid Midi Kit (Hilden, Germany). The plasmid was digested with BamHI and the DES3 fragment, withdrawn with Freeze 'N Squeeze DNA Gel Extraction Spin Columns, was ligated to pET15b expression vector pre-digested with the same enzyme to generate pET-DES3 recombinant plasmid.
CTATAGTTTCACATCTCTACC-3′; Tm 58 °C; position + 1206 respect to the start codon). Both primers were designed containing an adaptor with the BamHI restriction site. The length of the amplified product was 1203 bp. The recovery of the amplified product from agarose gel was obtained with Freeze 'N Squeeze DNA Gel Extraction Spin Columns made by Bio-Rad (Hercules, California). It was used pCR4-TOPO vector of 3956 bp (Invitrogen, Carlsbad, California) and Escherichia coli One Shot TOP10 Kit (Invitrogen, Carlsbad, California) to clone the DES3 sequence isolated as described above. The identity of the products was confirmed by sequencing. The isolation of plasmid pCR4-TOPO-DES3 DNA was obtained from QIAGEN Plasmid Midi Kit (Hilden, Germany). The plasmid was digested with BamHI and the DES3 fragment, withdrawn with Freeze 'N Squeeze DNA Gel Extraction Spin Columns, was ligated to pET15b expression vector pre-digested with the same enzyme to generate pET-DES3 recombinant plasmid.
The identity of the products was again confirmed by sequencing. The expression vector pET15b (5708 bp) was obtained from Novagen (Madison, Wisconsin). The pET-15b cloning/expression region carries a short N-terminal His-Tag sequence useful to purify the fusion protein obtained. After purification the His-Tag sequence can be removed using thrombin enzyme. In the obtained DES3 protein, the His-Tag sequence was not removed. E. coli BL21 Star (DE3) (Invitrogen) was used as the host for the expression of protein. Restriction enzyme (BamHI), T4 DNA Ligase, GoTaq® DNA Polymerase and Pfu DNA Polymerase were purchased from Promega (Madison, Wisconsin). All chemicals were products from Sigma-Aldrich unless otherwise specified.
2.2 Protein expression and analysis by SDS-PAGE
Single colony of recombinant E. coli BL21 strain carrying the vector pET-DES3 was added to 100 ml of LB medium containing 100 μg ml−1 ampicillin and cultured overnight at 37 °C with shaking.This culture was 20-fold diluted into fresh LB medium and grown two hours at 37 °C with shaking to an OD600 of about 0.5–0.8 (mid-log).
The culture was split into two 50 ml cultures and one of these was induced with isopropylthiogalactoside (IPTG) to a final concentration of 1 × 10−3 M.
The cultures were incubated to express the S-ACP-DES3 protein and cell growth was continued for 4 h. The cells were pelleted (1 ml) every hour, centrifuge at 12.000 g in a microcentrifuge for 1 minute at room temperature. The pellets were suspended in 100 ml of 1× SDS gel-loading buffer, heat to 100 °C for 3 minutes and then centrifuge; 15 μl of each suspension were loaded on a 10% SDS-polyacrilamide gel. Proteins were stained with Coomassie brilliant blue as described by Sambrook et al.19
2.3 Growth curve
E. coli BL21 strain carrying the vector pET-DES3 was added to 10 ml of LB medium containing 100 μg ml−1 ampicillin and cultured overnight at 37 °C with shaking. The culture was 50-fold diluted into fresh LB medium (1 ml of overnight culture to 50 ml of fresh LB medium) and grown at 37 °C with shaking measuring the OD600 of the culture every hour. LB broth was composed by 10 g l−1 Tryptone, 5 g l−1 Yeast Extract and 5 g l−1 NaCl (6.8–7.2 pH).2.4 Lipid extraction, FAME preparation and GC-MS analysis
Both control and the recombinant strain were propagated with 1× M9 minimal medium including 2% glucose as carbon source. About 400 mg of wet bacterial cells were harvested from 400 ml of fermentation culture induced with IPTG (0.84 g l−1 per OD600). Lipids were extracted from pellets following the method described by Cao et al.15 with some modifications. Pellets were re-suspended in 1 ml of distilled water and 10 ml of CHCl3/CH3OH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The solution was mixed for 5 min and left over-night. The chloroform phase was evaporated with nitrogen and 4 millilitres of 6% (w/v) NaOH in methanol/water (4
1). The solution was mixed for 5 min and left over-night. The chloroform phase was evaporated with nitrogen and 4 millilitres of 6% (w/v) NaOH in methanol/water (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added to the dried lipids and saponified in a 60 °C water bath for 3 h. The esterification was obtained using 4 ml of BF3/CH3OH (1
1) was added to the dried lipids and saponified in a 60 °C water bath for 3 h. The esterification was obtained using 4 ml of BF3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and the solution was heated for 30 min at 60 °C. The fatty acid methyl esters (FAMEs) generated were extracted twice with 5 ml of hexane.
4) and the solution was heated for 30 min at 60 °C. The fatty acid methyl esters (FAMEs) generated were extracted twice with 5 ml of hexane.
Molecules were separated using a capillary column AGILENT-5MS 30 m × 250 μm × 0.25 μm (ID). Carrier gas was helium at a flow rate of 1 ml min−1. One μl of sample was injected using CTC PAL into the GC injection port at 250 °C in splitless mode. The temperature program was set at 75 °C for 8 min, raised to 330 °C at a rate of 4 °C min−1 and the final temperature was maintained for 25 min. The transfer line to the mass spectrometer was maintained at 250 °C. The mass spectra were obtained by electronic impact at 70 eV, and collecting data at an m/z range of 40–550. The FAME concentration was calculated quantitatively, by direct comparison with the external standard peak area (Sigma Aldrich, 4-7801). The analysis was repeated three times.
3. Results and discussion
3.1 Cloning and expression of Arabidopsis DES3 gene in E. coli
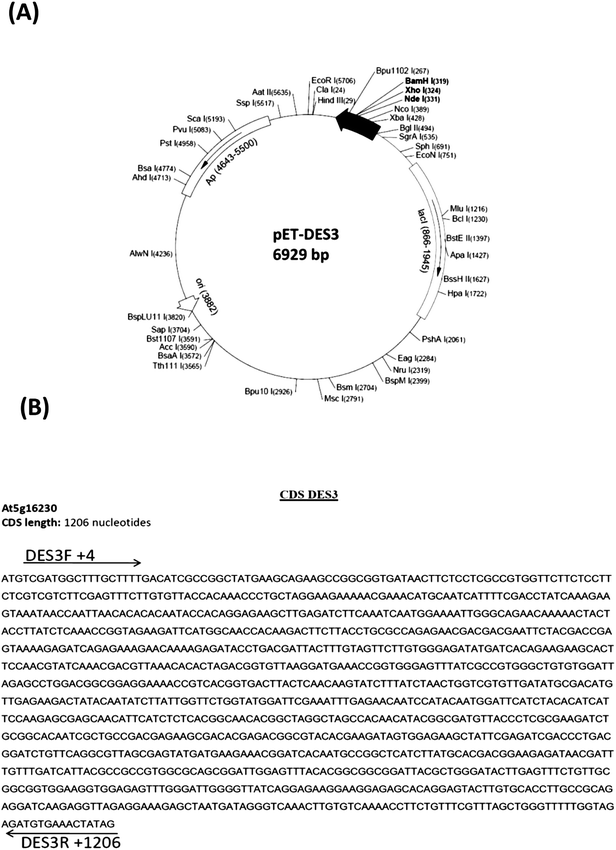
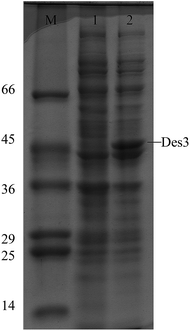
To express DES3 desaturase in E. coli the coding region of DES3 was cloned into pCR4-TOPO cloning vector (3956 bp) and then in the pET15b expression vector under the T7 promoter (Fig. 1) as described in the Material and methods chapter. The pET-15b vector allowed the transcription and translation of the sequence cloned after induction with IPTG (T7 promoter under control of Lac operator). This expression cannot be modulated, and the promoter works as an “on (+IPTG)/off (no IPTG)” switch. The expression construct, was checked by restriction enzyme digestion and DNA sequencing (data not shown). The recombinant plasmid was transformed into E. coli BL21 (DE3) strain and transformed cells with pET15b-DES3 were grown in sterilized liquid Luria-Bertani (LB) medium. This culture was diluted and split into two parts and one of these induced with 1 mM IPTG; both the cultures were incubated at 37 °C.In the SDS-PAGE analysis a strong band of about 46 kDa, i.e.DES3 protein in Arabidopsis thaliana, has been seen only in the extract from the pET-DES3 strain after protein induction. Contrarily band of 45 kDa was visible in both induced and non-induced protein extracts, demonstrating the correct expression in E. Coli of the DES3 protein, after 1 hour from IPTG induction (Fig. 2).
Furthermore the heterologous expression of Arabidopsis thaliana stearoyl-acyl carrier protein desaturase 3 (S-ACP-DES3), induced a slight reduction of the bacterial growth, compared to the not induced strain (Fig. 3). This decrease was not surprising, considering that the genetically modified bacteria was forced to produce a new protein able to modify the cell fatty acid composition15 (see the next paragraph).
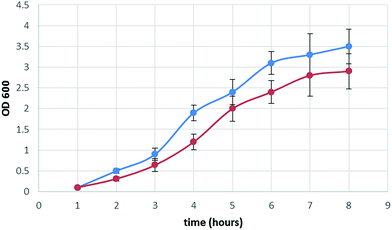 | ||
| Fig. 3 Growth curve of pET-DES3 strain (OD600) not induced (blue color) and of pET-DES3 strain after IPTG induction (red color). Error bars represent SD values (n = 3). | ||
3.2 Functional analysis of the DES3 protein in a heterologous system: change in fatty acid composition
With the aim of demonstrating the functionality of Arabidopsis DES3 protein in the E. coli organism, fatty acids profiles between the recombinant strain and the untransformed control were compared (Table 1).| E. coli BL21 control strain | E. coli recombinant pET-DES3 strain | ||
|---|---|---|---|
| mg g−1 dm pellet | |||
| a Values is significantly different to that for the E. coli BL21 control strain (p < 0.05), ANOVA bootstrap. | |||
| Dodecanoic acid methyl ester | C12:0 | 2.1 ± 0.1 | 3 ± 0.2 |
| Myristic acid methyl ester | C14:0 | 4.2 ± 0.1 | 6.8 ± 1.6 |
| Palmitic acid methyl ester | C16:0 | 32.8 ± 1.6 | 0.12 ± 0.01a |
| Palmitoleic acid methyl ester | C16:1 | 13.4 ± 0.3 | 13.3 ± 0.3 |
| Stearic acid methyl ester | C18:0 | 3.3 ± 0.1 | 0.02 ± 0.02a |
| Oleic acid methyl ester | C18:1 | 3.2 ± 0.1 | 31.9 ± 1.4a |
| Linoleic acid methyl ester | C18:2 | 4.6 ± 0.4 | 0.95 ± 0.26a |
| Linolenic acid methyl ester | C18:3 | 0.02 ± 0.01 | 0.54 ± 0.09a |
| cis-11 Eicosenoic acid methyl ester | C20:1 | 0.03 ± 0.01 | 0.13 ± 0.01a |
| cis-11,14-Eicosadienoic acid, methyl ester | C20:2 | 0.009 ± 0.002 | 0.94 ± 0.001a |
| 8,11,14-Eicosatrienoic acid methyl ester | C20:3 | 0.04 ± 0.01 | 0.65 ± 0.22a |
A total amount of 77.1 ± 5.6 mg g−1 dm pellet was found for E. coli before the transformation. Fatty acids were composed, above all, by palmitic acid (32.8 ± 1.6 mg g−1 dm pellet) and palmitoleic (13.4 ± 0.3 mg g−1 dm pellet) acids followed by myristic (4.2 ± 1 mg g−1 dm pellet), linoleic (4.6 ± 0.4 mg g−1 dm pellet), oleic (3.2 ± 0.4 mg g−1 dm pellet), stearic (3.3 ± 0.1 mg g−1 dm pellet) and dodecanoic (2.1 ± 0.1 mg g−1 dm pellet) methyl fatty acids. In addition some other FA were found at lower concentrations (Table 1).
The E. coli recombinant strain had a quite similar fatty acid content (58.5 ± 3.8 mg g−1 dm pellet) to the E. coli which was not transformed (77.1 ± 5.6 mg g−1 dm pellet), suggesting that this modification determined rather little changing of the total fatty acid production.
Total fatty acid contents before and after genetic transformation was of 6.7 ± 1.3% dm pellet (mean ± standard deviation) in line with literature that reported concentrations of 9.7% dm, for E. coli which had been genetically manipulated.20 However, due to the low amount of triacylglycerides production, the use of bacteria as raw source for biodiesel production is still nowadays restricted to laboratory scale.7 This is due to the fact that despite of easy genetic bacteria manipulability, in particular E. coli manipubility, microbial oil (named single cell oil – SCO) would seem, so far, more promising to obtain biodiesel thanks to their innate capacity to accumulate high levels of lipids rather than fatty acid quality. In particular microorganisms able to accumulate lipid above 20% (classified as oleaginous) have attracted the attention of the scientific community. For example the microalgaes Botryococcus braunii, Cylindrotheca sp. and Schizochytrium sp. accumulate oil respectively at 25–75, 16–37 and 50–77 as % of dry matter.7 Also some yeast strains are able to accumulate high level of lipid: Candida curvata, 58% dm, Rhodotorula glutinis 72% dm;7 nevertheless bacteria still remain a good candidate to became an important source of oil when genetic modifications will improve, also, oil content yields and growth rate.
Fatty acids composition greatly changed when the S-ACP-DES3 gene was expressed in E. coli (Table 1): SFA decreased from 72% to 17% of FA content while m-UFA increased from 21% FA to 77% FA.
These results demonstrate that the heterologous expression of S-ACP-DES3 in E. coli produced a functional protein that was able to change the balance in fatty acids desaturation with a specific effect regarding SFA and m-UFA. In particular big decreases of C16:0 (about 270 times) and of C18:0 (about 150 times) accompanied by an increasing of C18:1 (about 10 times) and others p-UFAs present in minor amounts were observed. No significant change in concentration occurred for C12:0 and C14 fatty acids (Table 1). Results obtained suggested that S-ACP-DES3 (At5g16230) acted mainly on C18:0 fatty acid, i.e. stearic acid desaturation producing the C18:1 oleic acid in agreement with the literature.16 It can be stated, also, that the decrease of C18:0 and C16:0 (a precursor of C18:0 in the fatty acid biosynthesis) was probably due to a down-regulation of the fatty acid pathway caused by an accumulation of C18:1. Hence this genetic manipulation altered not only the saturated/unsaturated fatty acids ratio but also the total balance of fatty acids composition: the ratio C18/C16 that in the untransformed E. coli was of 0.24 became in the transformed version of 2.50.
In order to evaluate the effect of the newly expressed gene on fatty acid composition and ultimately on biodiesel production, E. coli FAs profile was compared with those of vegetable oils, animal fats and of oleaginous microorganisms (microalgae, yeast and fungi) commonly used as oil-feedstocks to produce biodiesel.5,11,21 To do this the fatty acid composition (SFA < 26% FA, m-SFA > 62% FA, TFA < 7% FA; C20 + VLCFA < 19% FA) suggested by Moser and Vaughn,13 was adopted as terms for comparison (Fig. 4). None of the fats and oils considered matched the optimal composition indicated, apart from hazelnut and olive oils, which, however, are food-plants, producing 1st generation biodiesel. On the other hand modified E. coli (this work) showed an oil composition that perfectly respected the optimal fatty acid proportion, i.e. SFA 17% FA, m-SFA = 77.55% FA, TFA = 2% FA, C20 + VLCFA = 2.9% FA.
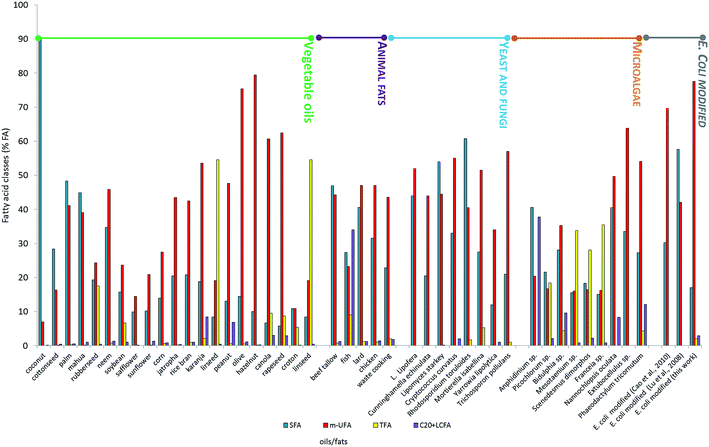 | ||
| Fig. 4 FAME composition of different biodiesel feedstocks (data from ref. 5, 7 and 9). | ||
4. Conclusion
This work shows that the Arabidopsis DES3 gene desaturase expressed into E. coli, modified fatty acid saturation giving optimal composition with regard to oil quality to be used as feedstock to produce biodiesel. These results can be used as a starting point to try to modify oleaginous bacteria, getting both high oil productivity and optimal oil composition.References
- E. Santacesaria, G. Martinez Vicente, M. Di Serio and R. Tesser, Catal. Today, 2012, 195, 1–168 CrossRef PubMed.
- R. Luque, L. Herrero-Davila, J. M. Campelo, J. H. Clark, J. M. Hidalgo, D. Luna, J. M. Marinas and A. A. Romero, Energy Environ. Sci., 2008, 1, 542–564 CAS.
- X. Meng, J. Yang, X. Xu, L. Zhang, Q. Nie and M. Xian, Renewable Energy, 2009, 34, 1–5 CrossRef CAS PubMed.
- H. J. Janßen and A. Steinbüchel, Biotechnol. Biofuels, 2014, 7, 1–26 CrossRef PubMed.
- G. Christophe, V. Kumar, R. Nouaille, G. Gaudet, P. Fontanille, A. Pandey, C. R. Soccol and C. Larroche, Braz. Arch. Biol. Technol., 2012, 1, 29–46 CrossRef PubMed.
- F. Adani, European Patent no. 2242555; F. Adani, US Pat. Application no. 12/866977, 2009.
- S. Pinzi, D. Leiva-Candia, I. López-García, M. D. Redel-Macías and M. Pilar Dorado, Biofuels, Bioprod. Biorefin., 2014, 126–143 CrossRef CAS.
- Q. Li, W. Du and D. Liu, Appl. Microbiol. Biotechnol., 2008, 80, 749–756 CrossRef CAS PubMed.
- M. A. Rude and A. Schirmer, Curr. Opin. Microbiol., 2009, 12, 274–281 CrossRef CAS PubMed.
- T. P. Durret, C. Benning and J. Ohlrogge, Plant J., 2008, 54, 593–607 CrossRef PubMed.
- E. G. Giakoumis, Renewable Energy, 2013, 50, 858–878 CrossRef CAS PubMed.
- S. K. Hoekman, A. Broch, C. Robbins, E. Ceniceros and M. Natarajan, Renewable Sustainable Energy Rev., 2012, 16, 143–169 CrossRef CAS PubMed.
- B. R. Moser and S. F. Vaughn, Biomass Bioenergy, 2012, 37, 31–41 CrossRef CAS PubMed.
- X. Lu, H. Vora and C. Khosla, Metab. Eng., 2008, 10, 333–339 CrossRef CAS PubMed.
- Y. Cao, J. Yang, M. Xian, X. Xu and W. Liu, Appl. Microbiol. Biotechnol., 2010, 87, 271–280 CrossRef CAS PubMed.
- E. B. Cahoon and J. Shanklin, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1235–1255 CrossRef PubMed.
- B. Behrouzian and P. H. Buist, Phytochem. Rev., 2003, 2, 103–111 CrossRef CAS.
- A. J. van Tunen, R. E. Koes, C. E. Spelt, A. R. van der Kroll, A. R. Stuitje and J. N. Mol, EMBO J., 1988, 7, 1257–1263 CAS.
- J. Sambrook, T. Maniatis and E. F. Fritsch, Molecular cloning : a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 3rd edn, 2001 Search PubMed.
- E. J. Steen, Y. Kang, G. Bokinsky, Z. Hu, A. Schirmer, A. McClure, S. B. del Cardayre and J. D. Keasling, Nature, 2010, 463, 559–562 CrossRef CAS PubMed.
- M. A. Islam, G. A. Ayoko, R. Brown, D. Stuart and K. Heimann, Procedia Eng., 2013, 56, 591–596 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |