Increased disulfide peptide sequence coverage via “cleavage ON/OFF” switch during nanoelectrospray†
Abstract
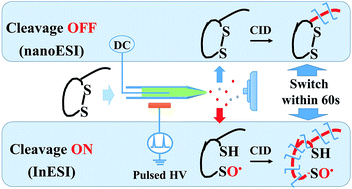
Herein, we developed an in situ disulfide cleavage protocol. It is based on the selective reaction between disulfide and the hydroxyl radical, which could be rapidly and controllably generated via induced nanoelectrospray. In less than one minute, increased disulfide peptide sequence coverage could be obtained via switching the “cleavage ON/OFF” modes between conventional and induced nanoelectrospray.

 Please wait while we load your content...
Please wait while we load your content...