Application of near infrared spectroscopy for the rapid determination of epimedin A, B, C and icariin in Epimedium
Abstract
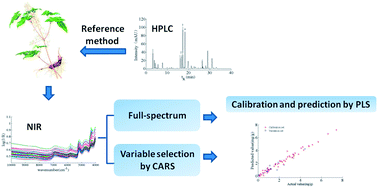
A method for rapid quantitative analysis of epimedin A, B, C and icariin in Epimedium was developed based on Fourier transform near infrared (FT-NIR) spectroscopy, and by adopting high performance liquid chromatography-diode array detection (HPLC-DAD) as the reference method. Multivariate calibrations models were built by partial least squares regression (PLSR) based on the full absorbance spectra (10 000–4000 cm−1) or only the most informative key variables selected by the competitive adaptive reweighted sampling (CARS) method. In comparison, the accuracy of the CARS-PLSR method was apparently higher than full spectrum-PLSR for four kinds of investigated flavonoids. For CARS-PLSR, the coefficients of determination (R2) for prediction were 0.8969, 0.8810, 0.9273 and 0.9325 and the root mean square errors of prediction (RMSEP) were 0.1789, 0.2572, 1.2872 and 0.3615 for epimedin A, B, C and icariin, respectively. The good performance indicates that the combination of NIR spectroscopy with CARS-PLSR is an effective method for determination of epimedin A, B, C and icariin in Epimedium with fast, economic and nondestructive advantages compared to traditional chemical methods.

 Please wait while we load your content...
Please wait while we load your content...