Coating of carboxymethyl dextran on liposomal curcumin to improve the anticancer activity†
Qiyu Huang,
Lili Zhang,
Xiaoyi Sun,
Ke Zeng,
Juan Li* and
You-Nian Liu*
College of Chemistry and Chemical Engineering, Central South University, Changsha, Hunan 410083, China. E-mail: juanli@csu.edu.cn; liuyounian@csu.edu.cn; Fax: +86-731-8887-9616; Tel: +86-731-8883-6964
First published on 3rd November 2014
Abstract
In this work, carboxymethyl dextran (CMD), a polysaccharidic dextran derivative with well-known biocompatibility and biodegradability, was used to modify curcumin-loaded liposomes for improving the anticancer efficacy of curcumin. The CMDylated curcumin-loaded liposomes (CMD-Cur-Lip) were successfully prepared and characterized in comparison with unmodified and two PEGylated curcumin-loaded liposomes. The results showed that: (1) the encapsulation efficiency of curcumin in the liposomes was improved after being modified with CMD; (2) all the curcumin-loaded liposomes displayed controlled release characteristics; (3) the CMD-Cur-Lip demonstrated improved cell cytotoxicity against cancer cells, as compared to other liposomal formulations and free curcumin; (4) CMD-Cur-Lip exhibited the highest cellular uptake as compared to other liposomal formulations.
Introduction
Curcumin is a natural polyphenol used for chemoprevention of multiple cancers by modulating multiple molecular pathways involving interference of tumor initiation, restriction of tumor progression, inhibition of invasion/metastasis of cancer cells and promotion of apoptosis.1 Nevertheless, the clinical application of curcumin is greatly hampered by its poor aqueous solubility therefore resulting in poor bioavailability.2 To overcome these limitations, nanoformulations of curcumin based on carriers such as biodegradable polymeric nanoparticles, micelles and liposomes, have been investigated.3Liposomes, due to their excellent biocompatibility and abilities to encapsulate and transport various molecules, are widely used in drug delivery, gene transfection and biosensing fields.4 Up to date, drug-carrying liposomes and modified liposomal nanoparticles are commercialized to treat certain cancers. For instance, PEGylated liposome-based doxorubicin (Doxil®) has been clinically approved in the treatment of ovarian cancer, AIDS-related Kaposi's sarcoma and multiple myeloma.5 Recently, liposomal curcumin has attracted a lot of attention. For example, curcumin-encapsulated liposome was found to suppress growth of head and neck squamous cell carcinoma both in vitro and in vivo via an AKT-independent pathway.6 Arab-Tehrany and coworkers reported a salmon lecithin based nanoliposomes for curcumin encapsulation to achieve improved curcumin bioavailability, as well as in vitro cytotoxicity through a real-time cell impedance monitoring.7
For prolonged blood circulation and increased cellular uptake, modifications of the liposomal curcumin were employed, among which PEGylation8 and chitosan coating9 are commonly used. Herein, we introduced an ameliorative modification to liposomal curcumin using carboxymethyl dextran (CMD). CMD, an anionic polysaccharidic derivative, possesses good biocompatibility, biodegradability, low immunogenicity and low toxicity.10 Our group has established an effective loading and controlled release of doxorubicin by CMD-coated liposomes.11 In this work, CMD was coated onto the liposomal curcumin through the attachment of a hydrophobic alkyl chain to CMD. Furthermore, PEGylated liposomes were also synthesized and compared with CMDylated liposomes in ways of encapsulation efficiency, curcumin release profile, cellular uptake and cell cytotoxicity.
Experimental
Materials
LIPOID E 80 (egg phospholipids with 80% phosphatidylcholine, agglomerates) was purchased from Shanghai Toshisun Biology & Technology Co., Ltd (Shanghai, China). Cholesterol was purchased from Sangon Biotech. (Shanghai, China). Bromoacetic acid was obtained from Bio Basic Inc. PEG10000 (MW 10000) and mPEG2000 (MW 2000) were purchased from Fluka. Oleylamine, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich. Curcumin (Cur, >98%) was purchased from Aladdin. Dextran-T10 of 10 kDa was purchased from Pharmacia AB (Uppsala, Sweden). Other materials were obtained from commercial sources.Synthesis of CMD–OA, mPEG–OA and PEG–OA
Detailed synthesis of all the compounds were described in ESI.† The synthesis of CMD–OA was similar to our previous work.11,12 Synthesis of mPEG–oleylamine (mPEG–OA) and PEG–oleylamine (PEG–OA) was based on Li and McLean's method.13 The contents of residual carboxyl groups in CMD–OA were measured by acid–base titration method, which were found to be 0.50 mmol [–COOH] g−1. The contents of carboxyl groups in PEG–OA were also measured by acid–base titration method, found to be 0.26 mmol [–COOH] g−1.Preparation of curcumin-loaded liposomes
In this work, four different curcumin-loaded liposomes, namely unmodified curcumin-loaded liposomes (Cur-Lip), PEGylated curcumin-loaded liposomes (PEG-Cur-Lip and mPEG-Cur-Lip) and CMDylated curcumin-loaded liposomes (CMD-Cur-Lip), were prepared according to the reported procedures.14 To be specific, exact 40 mg of LIPOID E 80, 5 mg of cholesterol, 2 mg of curcumin and 2 mg of modifying agent (PEG–OA, mPEG–OA and CMD–OA for PEG-Cur-Lip, mPEG-Cur-Lip and CMD-Cur-Lip, respectively) were weighed and dissolved in chloroform–methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) mixture solution. After sonication for 10 min, the organic solvents were removed by rotary evaporation, followed by 10 min N2 gas sweeping. The resultant yellow thin film was hydrated with 10 mL PBS (10 mM, pH 6.8) at 60 °C, with manual shaking until no obvious suspended particles were observed. Finally, the solution was sonicated to obtain the products. Blank liposomes were also prepared as described above except for the addition of curcumin.
1, v/v) mixture solution. After sonication for 10 min, the organic solvents were removed by rotary evaporation, followed by 10 min N2 gas sweeping. The resultant yellow thin film was hydrated with 10 mL PBS (10 mM, pH 6.8) at 60 °C, with manual shaking until no obvious suspended particles were observed. Finally, the solution was sonicated to obtain the products. Blank liposomes were also prepared as described above except for the addition of curcumin.
Encapsulation efficiency
The encapsulation efficiency (EE%) was measured by UV-Vis on a Shimadzu UV-2450. Curcumin-loaded liposomes were centrifuged at 7000 rpm for 5 min and 50 μL of supernatant were taken and dissolved in 2.5 mL of anhydrous ethanol. Absorbance at 425 nm was measured by UV-Vis and the concentration of loaded curcumin was calculated from the calibration curve. The encapsulation efficiency was obtained from the equation:| Encapsulation efficiency (%) = loaded curcumin/initial curcumin × 100 |
Calibration curve was obtained by plotting absorbance at 425 nm against curcumin concentrations of 2.0, 3.0, 4.0, 5.0, 6.0, 8.0 μg mL−1. Here, a linear equation was fitted as: A425 = 0.154C − 0.00204, R2 = 0.997.
Fluorescence spectroscopy
To verify the insertion of curcumin into the lipid bilayers, samples of curcumin-loaded liposomes were diluted with PBS (10 mM, pH 6.8) containing 0.5% (w/v) SDS to a final curcumin concentration of 2.44 μg mL−1, according to the encapsulation efficiency. SDS was added to increase the solubility of free curcumin in PBS and such concentration was used so that liposome stability would not be influenced by SDS.15 Free curcumin was dissolved in ethanol and then diluted by the above PBS/SDS into the same concentration as a control. The emission wavelength and fluorescence intensities of different samples were measured with excitation wavelength of 420 nm and scanning rate of 1200 nm min−1 (Hitachi, F-4600).Size distribution and ζ-potential measurement
The size distribution and ζ-potential was analyzed by dynamic light scattering (DLS) technique using a Malvern Zetasizer Nano ZS (Malvern instruments, UK). The apparatus was equipped with a 4 mW He/Ne laser emitting 633 nm. The hydrodynamic diameters (Dh), polydispersity index (PdI) and ζ-potential were measured at a scattering angle of 173° and 25 °C. All measurements were conducted in triplicates.Atomic force microscopy (AFM)
Morphological observation of CMD-Cur-Lip was performed on a Dimension V AFM instrument (Veeco, US). Sample was prepared by depositing 10 μL of diluted CMD-Cur-Lip (0.1 mg mL−1) on a silica chip and letting dry for 72 h at room temperature in a desiccator containing silica gel.In vitro release
The curcumin release profile from different liposomes was determined by dialysis method. The sample liposomes were centrifuged (7000 rpm, 5 min) to remove some possible insoluble curcumin and then 10 mL of the supernatant sample were dialyzed against 500 mL PBS/SDS solution. At certain time intervals, 3 mL of dialysis solution was taken out for analysis and fresh PBS/SDS solution was replenished. As for a complete curcumin release, dialysis solution would have a concentration of ∼4 μg mL−1 curcumin but its absorbance at 425 nm would be about 0.02 in PBS/SDS using a 1 cm pathlength cuvette, so it is necessary to modify the UV-Vis assay for curcumin quantification. In order to enhance the UV-Vis absorbance, equal volume of anhydrous ethanol (3 mL) was added into 3 mL of the dialysis sample since the extinction coefficiency for curcumin in PBS/SDS/ethanol is 10 times higher than that in PBS/SDS. Calibration curve was obtained by measuring A425 of free curcumin in PBS/SDS/ethanol and undergoing linear fitting. The cumulative release was calculated as follows:| Cumulative release (%) = released curcumin/total curcumin × 100 |
Fluorescence microscopy
HeLa cell lines were obtained from the cell bank of Xiangya Hospital, Central South University (Changsha, China). HeLa cell lines were grown in DMEM medium (GIBCO) containing 10% FBS, 1% streptomycin and 1% penicillin. The cells were maintained at 37 °C in a humidified incubator with 5% CO2. A 24-well plate was used and approximately 104 cells were seeded per well. When cell confluence reached ∼60%, they were ready to uptake the liposomes. Cur-Lip, PEG-Cur-Lip, mPEG-Cur-Lip and CMD-Cur-Lip were added into the wells with a final curcumin concentration of 20 μg mL−1 and incubated in the same condition for 4 and 24 h, respectively. Cellular uptake was analyzed by measuring fluorescence emission from curcumin using an inverted fluorescence microscope (Nikon ECLIPSE TE2000U) equipped with a digital camera and a blue filter (λex = 420–490 nm, λem = 490–560 nm). Images were captured with NIS Elements F 3.0 software (Nikon).Flow cytometry
To better characterize cellular uptake of the liposomal curcumin, we performed flow cytometry assay for quantification. HeLa cell lines were incubated similarly to the fluorescence microscopy, the difference being that approximately 2 × 105 cells were needed per well. After incubation with all four curcumin liposomes for 4 h at 37 °C in a humidified incubator with 5% CO2, cell culture was discarded and cells were digested by trypsin (GIBCO, 0.25%). Cells in each well were then harvested in 2 mL Eppendorf tubes and centrifuged at 8000 rpm for 5 min, followed by washing with 0.5 mL of 50 mM PBS (pH 7.4). Centrifugation and washing were repeated for three times and cells were finally suspended in 0.2 mL of PBS for flow cytometry measurement (BD, FACS 101). Untreated cells were used as blank control.MTT assay
MTT-based assay was conducted to assess the killing-effects of different liposomes against cancer cells. In general, HeLa cells were harvested and cultured until reaching logarithmic phase. The cells were then suspended and diluted with cell culture solution to reach a final cell concentration of approximately 1 × 105 per mL. A 96-well plate was prepared and 100 μL of cell suspension was added into each well. The cells were incubated in 37 °C thermal incubator containing 5% CO2 for 24 h. The culture solution was then discarded and each well was filled with 100 μL drug/liposomes (curcumin, Cur-Lip, PEG-Cur-Lip, mPEG-Cur-Lip and CMD-Cur-Lip) of different concentrations. Free curcumin was prepared by dissolving proper concentrations of free curcumin in DMSO. Incubation was performed for 48 h in the same condition. Twenty microliter of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in 2.5 mg mL−1 was added into each well and incubated for another 5 h. After the incubation, all the solutions in the wells were drawn and 200 μL DMSO was added into each well to dissolve the formed crystals. The plate was placed into a microplate reader (Bio-Tek ELx800) and the UV-Vis absorbance was determined at 490 nm (corrected for background absorbance at 630 nm).In addition, the data of cell viability for each concentration are statistically compared between CMD-Cur-Lip and the other four samples individually using the Student's t-test in Microsoft Excel 2007. The level of significance used is p < 0.05.
Results and discussion
Preparation and characterization of four liposomal curcumin
To increase the circulation of liposome, PEG coating is commonly utilized. Herein, CMD coating was applied to not only increase the circulation, more importantly, increase the cellular uptake of liposomes. In order to study the difference between CMD coating and traditional PEG coating, both CMDylated and PEGylated liposomes were fabricated. Specifically, PEG of MW 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and mPEG of MW 2000 were used as control.
000 and mPEG of MW 2000 were used as control.
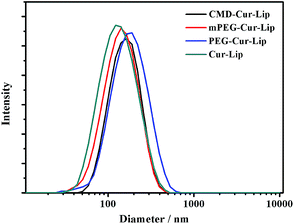
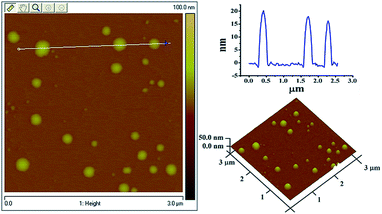
In order to coat mPEG, PEG and CMD onto the liposomal surface, chemical modification with lipid derivatives for an amphiphilic character was involved. After the EDC/NHS coupling, hydrophobic oleyl moieties were linked to the hydrophilic CMD and PEG molecules (ESI†). Then, the thin-lipid film hydration method was applied to load the hydrophobic curcumin into the liposomes. All four curcumin-loaded liposomes have average hydrodynamic diameters (Dh) of 120–200 nm (Fig. 1). As confirmed by AFM, the particles of CMD-Cur-Lip are spherical (Fig. 2). The size of liposomes has a range from 50 to 60 nm, with an average height of 10 nm. The size determined by AFM is much smaller than that by DLS, which might be caused by the different treatments during the tests. For AFM imaging, liposomal samples which were dried, could possibly collapse. On the other hand, the size distribution of liposomes by DLS was measured in a hydrated state.
After removing the unloaded curcumin precipitate from the liposomal dispersion by centrifugation, the EE% was calculated. Interestingly, the EE% of CMD-Cur-Lip and PEG-Cur-Lip are higher than those of PEG-Cur-Lip and unmodified Cur-Lip (listed in Table 1.). We also found higher EE% of CMD-Cur-Lip than those of other reported liposomal which were around 60–70%.7,16 The difference of EE% among the liposomes may be caused by the solubility of curcumin, and the stability of lipid components in the solution. The increase of EE% suggests CMD-Cur-Lip and PEG-Cur-Lip have higher stability than the others because the unloading of curcumin could influence the aggregation and/or fusion of liposomes. As reported, the solubility of curcumin in water is about 11 ng mL−1,17 the unloaded insoluble curcumin will precipitate in aqueous solution. As well, the liposomal suspension if not stabilized properly will undergo aggregation accompanying with insoluble curcumin. For CMD-Cur-Lip, CMD coating improves the stability of liposome and more curcumin could be well dispersed in the lipid layer, which is ascribed to the increase of EE%.
| Sample | EE/% | Dh/nm | PdI | ζ-potential/mV |
|---|---|---|---|---|
| Cur-Lip | 71.2 | 120.3 ± 6.2 | 0.19 ± 0.03 | −5.01 |
| mPEG-Cur-Lip | 77.7 | 139.4 ± 9.3 | 0.27 ± 0.02 | −5.04 |
| PEG-Cur-Lip | 81.5 | 184.1 ± 12.1 | 0.34 ± 0.05 | −7.63 |
| CMD-Cur-Lip | 82.8 | 141.5 ± 5.9 | 0.19 ± 0.02 | −8.58 |
Fluorescent properties of liposomal curcumin
It was reported by Puvvada that once curcumin was loaded into niosome, a nonionic surfactant-based vesicle, the maximum emission wavelength λmax would have a blue shift of around 50 nm due to its insertion into the lipid bilayer.18 This characteristic is an important indicator of curcumin loading and unloading. In our work, the fluorescence emission of curcumin did have a ∼30 nm shift after the curcumin encapsulation into the liposomes (Fig. 3). No obvious difference in emission wavelength was observed among the four liposomes, indicating that the liposomes probably encapsulated curcumin by the same mechanism of action. The blue shift suggests the presence of curcumin is transferred from the more polar region to liposomes containing less polar regions, consistent with other reported results.18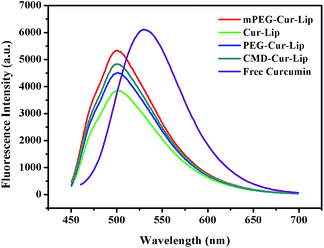 | ||
| Fig. 3 Comparison of fluorescence emission wavelengths from free and loaded curcumin under excitation of 420 nm. | ||
In vitro release
As discussed above, curcumin is hardly soluble in water. In addition, curcumin is extremely unstable when pH > 7.19 Due to the insolubility and instability of curcumin, the in vitro release experiment should be carefully carried out, especially for choosing the suitable release media, as also found by others.20 It is noted that the solubility and stability of curcumin could be improved in some surfactants, in cell culture medium containing 10% fetal calf serum and in human blood.20,21 In this work, the in vitro release of curcumin from the curcumin-loaded liposomes was measured in PBS/SDS (pH 6.8). PBS/SDS instead of PBS was used in order to keep the sink conditions which dissolved curcumin well. In addition, pH 6.8 was selected to lower the hydrolysis of curcumin. The results demonstrated that after 120 h of incubation, only 60% of the curcumin was released from CMD-Cur-Lip, 80–85% from mPEG-Cur-Lip and PEG-Cur-Lip, 90% from unmodified Cur-Lip (Fig. 4). While, most curcumin was released in the early 20 h for free curcumin. Generally, drug release is a complex process which is attributed to drug physicochemical properties, drug diffusion, and carrier integrity. The sustained release of CMD-Cur-Lip indicates the CMD coating stabilizes the liposomes against the leakage of curcumin in circulation. Also, this biocompatible CMD shell could reduce the risk of clearance by the reticuloendothelial system (RES),22 thus facilitating liposome accumulating at pathological sites via enhanced permeability and retention (EPR) effect.23 Furthermore, it was found that CMD-Cur-Lip are stable for at least 72 h upon incubation with calf serum (data not shown). It suggests that CMD-Cur-Lip are more potent than other three liposomes upon the systemic administration.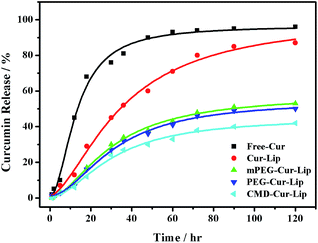 | ||
| Fig. 4 Cumulative release for curcumin alone and curcumin-loaded liposomes. Measurements were conducted in PBS/SDS(pH 6.8) at 37 °C for 120 h. | ||
Cellular uptake
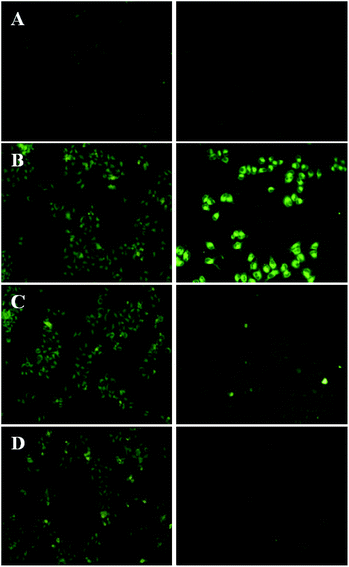
To observe the differences of liposomal curcumin in intracellular delivery, the uptake of Cur-Lip, CMD-Cur-Lip, PEG-Cur-Lip and mPEG-Cur-Lip by HeLa cells was determined by reverted fluorescence microscope, respectively. HeLa cells were treated with the same concentration of each curcumin-loaded liposomes, and the accumulation of curcumin in the cells was imaged after 4 h and 24 h, respectively. The results showed that after exposing for 1 s, cells incubated with all four liposomes generated decent fluorescence, among which unmodified Cur-Lip had a bit lowered fluorescent intensity compared with the other three (Fig. 5). This result clearly indicated the modification of either CMD or PEG helps the internalization of the liposomes into the cells. However, after 24 h incubation, even with increased exposure time (10 s), curcumin in the cells incubated with Cur-Lip became undetectable (Fig. 5A). PEGylated curcumin-loaded liposomes (mPEG-Cur-Lip and PEG-Cur-Lip) exhibited weakened curcumin accumulation (Fig. 5C and D), while CMD-Cur-Lip showed the strongest fluorescence intensity (Fig. 5B), expressing the greatest ability in delivering curcumin into HeLa cells and keeping the drug from leaking out of the cells, therefore inducing a prolonged and sustained drug treatment. The feature of enhanced intracellular uptake of CMDylated liposomes may be resulted from the facts that CMDylated liposomes selectively favor tumor cell-type and that the three-dimensional dextran hydroxyl network shell points to the endocytotic mechanisms of uptake.24To quantify the uptake efficiency of loaded curcumin into the cells, flow cytometry was conducted with excitation wavelength of 488 nm and emission wavelength of 520 nm. As seen in Fig. 6 untreated cells showed the weakest fluorescence emission with only 4.76% of cells gated in the positive zone. Cur-Lip, PEG-Cur-Lip and mPEG-Cur-Lip showed relatively stronger fluorescence intensity with 14.02%, 56.12% and 61.19% of cells gated, respectively. CMD-Cur-Lip generated the strongest fluorescence intensity with 92.99% of cells above the gate line, indicating the highest cellular uptake. Furthermore, the curcumin uptake amounts (calculated by the geometric mean of the fluorescence intensity) of CMD-Cur-Lip was about 3.7-fold higher than that of unmodified Cur-Lip, and about two-fold over those of mPEG-Cur-Lip and PEG-Cur-Lip. These results quantitatively supported the above visualization of cellular uptake by inverted fluorescence microscope, being a strong proof of enhanced cellular uptake by CMD coating. The enhanced cellular uptake of curcumin could be favorable for their anticancer activity, as demonstrated by others and our previous studies.25
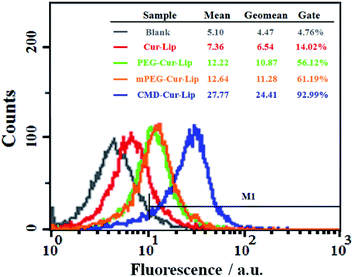 | ||
| Fig. 6 Flow cytometric measurement of curcumin uptake by HeLa cells after incubation with Cur-Lip, PEG-Cur-Lip, mPEG-Cur-Lip and CMD-Cur-Lip. | ||
Cell cytotoxicity
To determine whether the encapsulation of curcumin into modified or unmodified liposomes could enhance the anticancer activity, cell viability of HeLa cells were analyzed after curcumin or curcumin-loaded liposomes treatment. The results showed that liposomal encapsulation enhanced the cytotoxicity of curcumin towards the cancer cells (Fig. 7). With the increase of curcumin concentration, all groups showed higher cytotoxicities, among which CMD-Cur-Lip exhibited the strongest cytotoxicity. To be exact, the IC50 of Cur-Lip was ∼15.6 μM. Treatments with PEG-Cur-Lip, mPEG-Cur-Lip and CMD-Cur-Lip led to a lowered IC50 of ∼13.1 μM, ∼11.4 μM and ∼6.6 μM, respectively, showing a greatly improved cytotoxicity as compared with free curcumin (IC50: ∼24.8 μM). As derived from the statistical analysis, there are significant differences between cytotoxicity of CMD-Cur-Lip and those of other three curcumin-loaded liposomes and free curcumin at the high curcumin concentrations (20 μM and 50 μM). Therefore, CMD-Cur-Lip significantly enhanced the inhibition of tumor cell growth. To further support our findings, the cytotoxicity of blank liposomes (Lip, PEG-Lip, mPEG-Lip and CMD-Lip) were also tested (Fig. 8). The stock solutions of blank liposomes were prepared exactly according to the procedure of curcumin-loaded liposomes, except for the absence of curcumin. The stock solution was then serial-diluted in the same multiples so that the lipid concentration of blank liposomes remained the same as used in the previous MTT assay. It was revealed that no significant differences among the cytotoxicity of the blank liposomes at all the concentrations studied. This result strongly suggests cytotoxicity of CMD-Cur-Lips shown in the MTT assay was the outcome of the enhanced curcumin intracellular uptake by the cancer cells.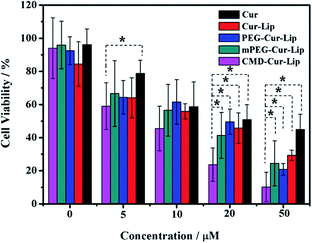 | ||
| Fig. 7 Cell viability of HeLa cells as a function of curcumin concentration. Asterisks indicate significant differences (p < 0.05). | ||
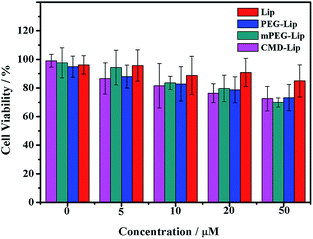 | ||
| Fig. 8 Cell viability of HeLa cells treated with blank liposomes as a function of lipid concentration. Axis of coordinates herein is expressed in the values of curcumin concentration in Fig. 7 for comparing with the corresponding cytotoxicity of control liposomal curcumin. | ||
Conclusion
In brief, this work has shown that CMD coating on liposomal curcumin improves the cytotoxicity of curcumin against cancer cells by enhancing the sustained release and cellular uptake. Moreover, the residual carboxyl groups on the CMD layer also allow for further modifications such as targeting ligands. These characteristics render the CMD-Lip a potential vector for the encapsulation of other hydrophobic antitumor drugs, with which further studies are underway.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21374133, 21276285 and 21104096) and Hunan Provincial Natural Science Foundation of China (no. 12JJ4014).Notes and references
- (a) S. S. Dhule, P. Penfornis, J. B. He, M. R. Harris, T. Terry, V. John and R. Pochampally, Mol. Pharm., 2014, 11, 417 CrossRef CAS PubMed; (b) W. Park, A. Amin, Z. G. Chen and D. M. Shin, Cancer Prev. Res., 2013, 6, 387 CrossRef CAS PubMed; (c) R. L. Thangapazham, A. Sharma and R. K. Maheshwari, AAPS J., 2006, 8, E443 CrossRef CAS PubMed.
- S. Jambhrunkar, S. Karmakar, A. Popat, M. H. Yu and C. Z. Yu, RSC Adv., 2014, 4, 709 RSC.
- (a) M. Salem, S. Rohani and E. R. Gillies, RSC Adv., 2014, 4, 10815 RSC; (b) S. Barui, S. Saha, G. Mondal, S. Haseena and A. Chaudhuri, Biomaterials, 2014, 35, 1643 CrossRef CAS PubMed; (c) M. M. Yallapu, M. Jaggi and S. C. Chauhan, Drug Discovery Today, 2012, 17, 71 CrossRef CAS PubMed.
- Q. Liu and B. J. Boyd, Analyst, 2013, 138, 391 RSC.
- Y. Barenholz, J. Controlled Release, 2012, 160, 117 CrossRef CAS PubMed.
- D. Wang, M. S. Veena, K. Stevenson, C. Tang, B. Ho, J. D. Suh, V. M. Duarte, K. F. Faull, K. Mehta, E. S. Srivatsan and M. B. Wang, Clin. Cancer Res., 2008, 14, 6228 CrossRef CAS PubMed.
- M. Hasan, N. Belhaj, H. Benachour, M. Barberi-Heyob, C. J. F. Kahn, E. Jabbari, M. Linder and E. Arab-Tehrany, Int. J. Pharm., 2014, 461, 519 CrossRef CAS PubMed.
- (a) J. Y. Liu, S. Y. Chen, L. Lv, L. Song, S. R. Guo and S. T. Huang, Curr. Pharm. Des., 2013, 19, 1974 CAS; (b) Y. L. Lin, Y. K. Liu, N. M. Tsai, J. H. Hsieh, C. H. Chen, C. M. Lin and K. W. Liao, Nanomedicine, 2012, 8, 318 CrossRef CAS PubMed.
- (a) A. Karewicz, D. Bielska, A. Loboda, B. Gzyl-Malcher, J. Bednar, A. Jozkowicz, J. Dulak and M. Nowakowska, Colloids Surf., B, 2013, 109, 307 CrossRef CAS PubMed; (b) G. H. Shin, S. K. Chung, J. T. Kim, H. J. Joung and H. J. Park, J. Agric. Food Chem., 2013, 61, 11119 CrossRef CAS PubMed.
- (a) E. Antonini, L. Bellelli, M. L. Bonacci, M. R. Bruzzesi, A. Caputo, E. Chiancone and A. Rossi-Fanelli, Biopolymers, 1964, 2, 27 CrossRef CAS; (b) E. Antonini, L. Bellelli, M. L. Bonacci, M. R. Bruzzesi, A. Caputo, E. Chiancone and A. Rossi-Fanelli, Biopolymers, 1964, 2, 35 CrossRef CAS.
- S. Ning, Q. Huang, X. Sun, C. Li, Y. Zhang, J. Li and Y.-N. Liu, Soft Matter, 2011, 7, 9394 RSC.
- K. Wagner, A. Kautz, M. Roder, M. Schwalbe, K. Pachmann, J. H. Clement and M. Schnabelrauch, Appl. Organomet. Chem., 2004, 18, 514 CrossRef CAS.
- (a) X. Q. Li, F. T. Meng, H. Guang and Z. G. Su, J. Chem. Res., 2005, 2005, 280 CrossRef; (b) K. M. McLean, G. Johnson, R. C. Chatelier, G. J. Beumer, J. G. Steele and H. J. Griesser, Colloids Surf., B, 2000, 18, 221 CrossRef CAS.
- (a) S. S. Dhule, P. Penfornis, T. Frazier, R. Walker, J. Feldman, G. Tan, J. He, A. Alb, V. John and R. Prchampally, Nanomedicine, 2012, 8, 440 CrossRef CAS PubMed; (b) Y. M. Niu, X. Y. Wang, S. H. Chai, Z. Y. Chen, X. Q. An and W. G. Shen, J. Agric. Food Chem., 2012, 60, 1865 CrossRef CAS PubMed.
- H. Wang, P. Li, L. Zhang, J. Zhang, X. Ling and H. Ran, China J. Chin. Mater. Med., 2009, 34, 3021 CAS.
- M. Takahashi, S. Uechi, K. Takara, Y. Asikin and K. Wada, J. Agric. Food Chem., 2009, 57, 9141 CrossRef CAS PubMed.
- Y. Kaminaga, A. Nagatsu, T. Akiyama, N. Sugimoto, T. Yamazaki, T. Maitani and H. Mizukami, FEBS Lett., 2003, 555, 311 CrossRef CAS PubMed.
- N. Puvvada, S. Rajput, B. N. P. Kumar, M. Mandal and A. Pathak, RSC Adv., 2013, 3, 2553 RSC.
- H. Tonnesen and J. Karlsen, Z. Lebensm.-Unters. Forsch., 1985, 180, 402–404 CrossRef CAS.
- L. Song, Y. Y. Shen, J. W. Hou, L. Lei, S. R. Guo and C. Y. Qian, Colloids Surf., A, 2011, 390, 25–32 CrossRef CAS.
- Y. Wang, M. Pan, A. Cheng, L. Lin, Y. Ho, C. Hsieh and J. Lin, J. Pharm. Biomed. Anal., 1997, 15, 1867–1876 CrossRef CAS PubMed.
- M. Hashida, M. Nishikawa, F. Yamashita and Y. Takakura, Drug Dev. Ind. Pharm., 1994, 20, 581–590 CrossRef CAS.
- R. R. Sawant and V. P. Torchilin, Soft Matter, 2010, 6, 4026 RSC.
- M. Schwalbe, N. Buske, M. Vetterlein, K. Hoffken, K. Pachmann and J. H. Clement, Z. Phys. Chem., 2006, 220, 125 CrossRef CAS.
- L. Zhang, Z. Q. Qi, Q. Huang, K. Zeng, X. Sun, J. Li and Y.-N. Liu, Colloids Surf., B, 2014, 123, 15 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of CMD–OA, mPEG–OA, and PEG–OA. See DOI: 10.1039/c4ra11181h |
| This journal is © The Royal Society of Chemistry 2014 |