Catalytic inactivation of alkaline phosphatase by cantharidin, an inhibitor of protein phosphatase
Rashid Ahmed Khan†
,
Jiyuan Liu† and
Yalin Zhang*
Key Laboratory of Plant Protection Resources and Pest Management, Ministry of Education, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, P.R. China. E-mail: yalinzh@nwsuaf.edu.cn; Fax: +86-29-87092190; Tel: +86-29-87092190
First published on 1st October 2014
Abstract
Previous investigations have shown high toxicity of cantharidin to many insects especially lepidopteran. However, its use in a higher dose for pest management has raised serious environmental concerns. Therefore, its biological potential in sub-lethal dose as a synergist was considered. It is essential for a synergist to be an effective inhibitor of metabolic enzymes, especially those responsible for biotic and abiotic stresses. As alkaline phosphatases are an important type of enzyme involved in numerous physiological processes and also in insecticide resistance, any impairment in their function may lead to serious physiological disturbances and could compromise their catalytic activity. Results showed that a sub-lethal dose of 25 μg g−1 treated artificial diet fed to Helicoverpa armigera showed inhibitory effects on catalytic activity of alkaline phosphatase in the insect midgut. Furthermore, kinetic data showed that cantharidin inhibited HaALPs competitively with respect to p-NPP. The inhibitory effect of cantharidin on the catalytic activity of ALPs as a result of its binding to the putative catalytic site was also confirmed by using homology modelling, molecular dynamics and docking simulations.
Alkaline phosphatase (ALP) (EC 3.1.3.1) is an important metalloenzyme that is responsible for the removal of phosphate groups from organic compounds. ALPs show similar properties in hydrolase, transferase reactions. Their activity is dependent on Zn2+ and Mg2+ ions and sequences of identical amino acids around an active site.1 ALPs are hydrolytic in nature and their function is to hydrolyze phosphomonoesters under alkaline conditions. ALPs are mainly found in the intestinal epithelium of animals and their primary role is to provide phosphate ions from mononucleotide and ribonucleo-proteins for diverse metabolic processes. ALPs have been found to be involved in the transphosphorylation reaction.2 Any impairment or interruption in their function will affect the physiology of insects, especially their gut. Enzymes belonging to this family are found mainly in muscle, nerve fibers, midgut and malpighian tubules of the lepidopteran insects.3 As compared to other tissues, the midgut has the highest ALP activity. The widespread presence of ALPs suggests their involvement in numerous biochemical functions, however there is no definitive proof about their exact physiological functions or knowledge of their natural substrate.4 Protein phosphatase activity and involvement in the proliferation of cells are also regarded as possible functions of ALP.5
In addition, ALPs have been found to involve in several physiological processes and respond to biotic and abiotic stress.1,6–9 The role of the ALP is crucial for the synthesis of tyrosine and act as a precursor of dopamine and octopamine, which are responsible for control of levels of insect developmental hormones such as, juvenile and 20-hydroxyecdysone hormones.10–12
Although, there is no direct evidence of ALP involvement in insecticide metabolism but some evidences suggest involvement of ALPs in insecticide resistance. For instance, higher level of ALPs in the resistant strain compared to susceptible strain was reported earlier in fenvalerate resistant population of Helicoverpa armigera.13 Furthermore, application of methomyl and parathion also caused increased ALP activity in H. armigera.14
Cantharidin found in blister beetles is a well-known natural product derived from insects.15–18 Cantharidin as an emulsifiable concentrate (1% EC) has been used in China as a bio-pesticide.19 Its antifeedant and insecticidal activity to pests have been demonstrated earlier.20 However, its exact mechanism of toxicity to insects is not known. Earlier studies showed bioactivity of cantharidin is due to its binding to phosphoprotein 2A (PP2A-AC).21 Other than its binding to PP2A, detailed physiological and biochemical effects and its mode of action is widely unknown.22–26 In the previous investigation alkaline phosphatase from Escherichia coli type III-S, placental ALP and calf intestinal ALP were inhibited by okadiac acid (OA) an inhibitor of protein phosphatases PP1 and PP2A.4 Among insects, cantharidin has been reported earlier for its inhibitory effects on alkaline phosphatase in armyworm, Mythimna separate, in vivo.27 The role of cantharidin as a synergist was also explored for diamondback moth, Plutella xylostella using cantharidin with other insecticides, namely, abamectin, endosulfan, chlorfluazuron, bisultap and methomyl. Among the mixtures cantharidin mixture with chlorfluazuron showed the highest synergism and was regarded as the best mixture.28 However, previous investigations lacked detailed studies at molecular, biochemical and computational level.
Present investigations were carried out to test the inhibitory effect of cantharidin on catalytic activity of ALP in the midgut of H. armigera, in view of the preliminary report on inhibition of ALP by cantharidin and its putative protein phosphatase activity, using, enzyme kinetic and molecular dynamic and docking simulations. For this reason enzyme kinetic studies were carried out in vitro to investigate the type of inhibition. In addition, binding of cantharidin to the putative catalytic active site of HaALP was investigated using homology modelling and molecular dynamics and docking simulations.
Results and discussion
Specific activity of HaALP in vivo
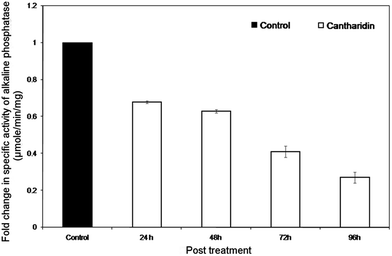
The inhibitory effects of cantharidin on HaALP enzyme within midguts dissected from larvae treated with sub-lethal dose of cantharidin is shown in Fig. 1. A sub-lethal dose of 25 μg g−1 exerted inhibitory effects on HaALP in the midgut compared to untreated control. Data showed that the specific activity of HaALP tended to decrease in post treatment from 24 to 96 h, whereas specific activity tended to increase in untreated controls. The inhibitory effect on the activity at 96 h after treatment was 22 compared to 82 μmol min−1 mg−1 of control.Kinetic properties of ALP
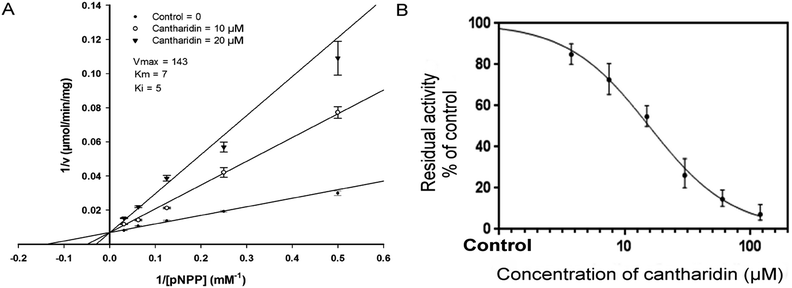
To find out the type of inhibition of HaALP enzyme extract by cantharidin, HaALP activity was calculated with variable concentrations of p-NPP. Lineweaver–Burk plot for p-NPP as the variable substrate and kind of inhibition by cantharidin is shown in (Fig. 2A). Lineweaver–Burk plot showed that cantharidin inhibited HaALP competitively as Vmax remained unchanged while Km decreased with respect to p-NPP. To calculate IC50 of cantharidin for HaALP, variable concentrations of cantharidin were used using p-NPP as substrate. Results showed that cantharidin inhibited HaALP in a dose-dependent manner with 50% inhibitory concentration of 42.95 μM (Fig. 2B).Homology modelling
HaALP sequence alignment with a PDB sequence of 1K7H X-ray crystallographic structure at the resolution of 1.92 Å was found to have the consensus sequence of 43% with 88% coverage (Fig. 3). Shrimp Alkaline Phosphatase (SAP; 1K7H) was found suitable to act as template of HaALP. Ramachandran plot showed that the HaALP main chain structure was reasonable. Amino acid residues located in the optimum area were 97.4%, whereas 98.5% residues located in the acceptable region. Six amino acid residues HIS99, SER253, LEU294, PRO 295, ARG412 and PRO447 were found far away from the active site. | ||
| Fig. 3 Multiple sequence alignment. The target protein, HaALP (ACF40806) and template protein, 1K7H sequences. The deep green color shows conserved amino acid residues in both the sequences. | ||
Molecular dynamics simulations
The RMSD curve became flat after 2 ns, indicating the conformations of the protein reached equilibrium. The average Cα RMSD value (from 2 ns to the end) for the ensemble was 2.2 ± 0.1 Å. The RMSD plots indicated that the conformations of HaALP ensemble achieve equilibrium, suggesting that this conformation is stable. Average conformation was applied as the basis for molecular docking conformation after 5 ns equilibration (Fig. 4). | ||
| Fig. 4 RMSD of the Cα atoms of the HaALP. The plot showing line became flat after 5 ns representing stability of the structure. | ||
3-D model structure superimposition
HaALP was found having ten-stranded β-sheets, sandwiched between a set of helices of various lengths, three on one side and five on the other side of the sheet. The b-strands are parallel, except for one anti-parallel insertion (Fig. 5A). Superimposition of the Cα backbone of HaALP with SAP crystal structure (RMSD value 0.132) showed similar topological characteristics (Fig. 5B). | ||
| Fig. 5 (A) Ribbon model of HaALP. HaALP is having ten-stranded β-sheets, sandwiched between a set of helices of various lengths, three on one side and five on the other side of the sheet. The b-strands are parallel, except for one anti-parallel insertion. (B) Superposition of the modelled structure of HaALP (Gray) with the X-ray crystal structure of 1K7H (Blue). Only backbones are shown in the picture. (C) The surface model of cantharidin with the HaALP active site. (D) The contact residues of cantharidin with HaALP. The contact residues here refer to the residues that are within 4 Å of cantharidin atoms. (E) Interaction of cantharidin with SER491 (F) Interaction of cantharidin with VAL114 (G) interaction of cantharidin with TYR112. (H) Interaction of cantharidin with LYS133. These interactions were generated by ligplot. | ||
Binding model analysis
After the docking calculation by AutoDock, four better binding models were obtained. Rank 1 was selected based on lowest docking energy and clustering circumstances (Table 1). Analysis of the structure of binding complex, cantharidin–HaALP as shown in (Fig. 5C) showed that cantharidin docked completely in the active centre of HaALP. Contact residues between the receptor and ligand are shown in (Fig. 5D). Ligplot package was utilized to compute the interaction diagram (Fig. 5E, F, G and H) among the cantharidin and the key amino acids; SER491, TYR112 VAL114, and LYS133. Hydrogen bonds play important roles in the structure and function of the macromolecule, especially for the catalysis of the enzyme. The HB plot analysis29 showed that four hydrogen bonds interactions were formed among cantharidin and amino acid residues TRY112, VAL114, LYS133 and SER491 of HaALP active site. OH of TRY112 and NZ of LYS133 both is hydrogen bond donor for cantharidin, whereas O of TRY112 is a hydrogen bond acceptor for LYS133. The distance of NZ and O is 2.65 Å. N of VAL114 is also a hydrogen bond donor for cantharidin. With the help of these hydrogen bonds cantharidin may occupy the active site and form stable complexes with HaALP and may mask the catalytic activity of Zn2+ in the active centre of HaALP.| Rank | Free energy of binding (kcal mol−1) | Inhibition constant, Ki (μM) | vdW + H bond + dissolve energy (kcal mol−1) | Electrostatic energy (kcal mol−1) | Total intermolec. energy (kcal mol−1) | Frequency | Interact. surface (Å2) |
|---|---|---|---|---|---|---|---|
| 1 | −2.0 | 31.7 | −2.6 | +0.60 | −2.0 | 60% | 315.8 |
| 2 | −2.0 | 32.2 | −3.0 | +1.01 | −2.0 | 10% | 323.6 |
| 3 | −1.8 | 45.4 | −2.1 | +0.33 | −1.8 | 10% | 270.1 |
| 4 | −1.7 | 48.4 | −2.4 | +0.65 | −1.7 | 20% | 295.4 |
In an earlier investigation toxicity of cantharidin and its analogues was attributed to its high affinity and specificity for cantharidin binding protein (CBP). Based on the similarities of four amino acid peptides from heterodimeric CBP α and β with Aα regulatory and Cβ catalytic subunits of protein phosphatase 2A (PP2A), CBP was assigned as PP2A-AC.21 Other than its high affinity to PP2A-AC exact physiological, biochemical effects and mode of action is widely unknown.22–26 In the same study okadaic acid (OA) competitively inhibited binding of cantharidin to pure CBP showing similar binding site. In the present study we chose cantharidin as an inhibitor of ALP based on the previous investigation showing OA a protein phosphatase inhibitor as an inhibitor of the catalytic activity of alkaline phosphatase from E. coli, human placental and calf intestinal ALP.
In the present investigation, cantharidin in sub-lethal concentration of 25 μg g−1 exerted inhibitory effects on the catalytic activity of ALP in the midgut. Moreover, the double reciprocal plot showed cantharidin competitively inhibiting catalytic activity with respect to p-NPP. Our results are in line with the previous investigation showing OA inhibiting catalytic activity of ALP.
Earlier cantharidin was found to be very toxic to Mythimna separata in higher concentration.27 Moreover, its sub-lethal concentration was used for the determination of its inhibitory effects on catalytic activity of metabolizing enzymes including ALP in vivo. The results showed inhibition of catalytic activity by cantharidin. In another study cantharidin was used as synergist with other insecticide and results showed a high level of synergism by cantharidin indicating its interaction with more than one mechanism for its toxicity to insects.28
At present, no published data are available about the inhibition of ALPs by cantharidin from other organisms. We believe detailed studies are required to investigate the inhibition of ALPs from other organisms, such as mammals, bacteria, fungi etc. for better understanding of the inhibitory mechanism. Moreover, homology modelling and molecular docking simulation techniques may be employed for sequence and conformational similarities of ALPs from other organisms and their role in protein ligand interaction.
Inhibition of ALP by cantharidin suggests the possibility of its involvement in the phosphorylation process by regulating phosphorylation/dephosphorylation of the membrane bound protein.4 Protein phosphatase activity of dephosphorylating proteins with phosphorylated tyrosine residues has been shown by alkaline phosphatise.30 However, cantharidin has not shown inhibitory effects on tyrosine phosphate and tyrosine kinases but is regarded as a selective inhibitor of protein serine threonine phosphatases.31–34 Consequently, we may conclude that inhibition of ALP by cantharidin suggests its role in phosphoseryl and phosphothreonyl phosphatase activity.4 A specific PHO13 ALP from Saccharomyces cerevisiae was found to exhibit protein phosphatase activity on the phosphoseryl protein histone II-A and casein.35
Furthermore, the molecular docking approach was used in order to understand the interaction of cantharidin to the putative catalytic active site. At first structures of two homologous ALPs, HaALP and SAP were modelled and optimized successfully. At present homology modelling can effectively be used for prediction of the structure of a particular protein. The power of this technique is attributed to the sequence identity between the template and target structure. The structure with more identity is considered better. Although the modelling of our target protein with crystal structure of SAP indicating the identity of 43% but, their secondary and tertiary structures are highly conserved. Superimposition of HaALP and SAP crystal structures revealed closest homology within Cα backbones having RMSD of 0.064 Å. The suitability of the structure was validated using Ramachandran plot. More than 90% of the amino acid residues were found in 94.7%, while 98.5% was found in acceptable region. Based on the results of superimposition and Ramachandran plot showed that our optimized model is suitable for reliable molecular docking.
The optimum docking conformation was selected based on lowest binding free energy for docking and most cluster members were selected after binding modes clustering through root mean square deviation (RMSD). The better binding model is attributed to the minimum energy for the formation of a complex between ligand and receptor.36 The formation of stable complex between ligand and receptor through hydrogen bonds within Zn2+ containing active cavity of HaALP suggested the unavailability of the active centre for the its catalytic activities or making it inactive by conformational changes.
Materials and methods
Test Insect
Cotton bollworm, Helicoverpa armigera larvae were procured commercially from Beijing Zhongke Baiyun Biotech. Co., Ltd. China and were reared under laboratory conditions using artificial diet.37 Batches of 24 larvae were placed into 24-chamber plastic boxes. The boxes were placed in an incubator at 27 + 1 °C and 40 to 50% RH with a 12 h photoperiod.Insect treatment
Cantharidin was extracted and purified38 in the laboratory. An earlier experimentally determined sub-lethal concentration of 25 μg g−1 cantharidin was used to investigate its effects on gene regulation of HaALP and catalytic activity of ALPs, using a diet incorporation bioassay. Third instar larvae were kept hungry for 8 h before their introduction to cantharidin-treated diet in a concentration of 25 μg g−1. The larvae were collected at 24, 48, 72 and 96 h and flash frozen in liquid nitrogen for subsequent extraction of total mRNA and protein. Ten larvae were used per replication and three biological replicates were used for all the treatments.Enzyme preparation and alkaline phosphatase activity estimation assay
A batch of 30 larvae each from treatment and control groups was collected randomly at 24, 48, 72 and 96 h. Collected insects were flash frozen in liquid nitrogen and stored at −80 °C in freezer. Midguts were dissected from larvae stored at −80 °C and washed with ice-cold 1× phosphate buffer. Homogenates were prepared by homogenizing midguts with glass homogenizer in ALP buffer containing 0.824 g sodium barbital, 0.35 mL of 0.2 M HCl mixed in 100 mL of water. The homogenates were subjected to centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min at 4 °C. The supernatants were collected as enzyme source. In addition, the supernatant extract was desalted using Sephadex G-25 spin columns to remove the contaminating free phosphate.
000 × g for 15 min at 4 °C. The supernatants were collected as enzyme source. In addition, the supernatant extract was desalted using Sephadex G-25 spin columns to remove the contaminating free phosphate.
To test alkaline phosphatase inhibition in the midgut from larvae treated with cantharidin, an assay was performed with some modifications.39 Preliminary experiments were carried out to further standardize the alkaline phosphatise assay. Special attention was given to maintain pH 9.6 of the reaction mixture. The experiment was replicated and in case of higher deviations the data were discarded. The alkaline phosphatase assay was carried out in 96 wells micro-plate. The total assay volume of 200 μL containing 130 μL of assay buffer, 10 μL of 4× diluted crude enzyme extract and 10 μL of 10 mM p-NPP solution, was used. The reaction was carried out at 37 °C for 30 min. The reaction was stopped by adding 50 μL 0.5 N NaOH. The hydrolysis of p-NPP to p-NP was recorded by measuring absorbance at 405 nm using TECAN™ Infinite® 200 PRO multimode micro-plate reader.
Enzyme kinetics and inhibition study
Kinetic study of enzyme inhibition was carried out using different concentrations of p-NPP and two concentrations of cantharidin, 10 and 20 μM. For the assay enzyme was extracted as mentioned above. The extracted enzyme was diluted 4× to keep the absorbance below 0.8 at 405 nm. The alkaline phosphatase kinetic assay was carried out in 96 wells micro-plate. The total assay volume of 200 μL contained 170 μL assay buffer, 10 μL of 10× diluted enzyme extract, 10 μL of 10 and 20 μM cantharidin in acetone and variable concentration of p-NPP. The reaction was pre-warmed for 10 min at 37 °C before adding p-NPP substrate. The hydrolysis of p-NPP to p-NP was recorded by measuring absorbance at 405 nm. Enzyme kinetics module of Sigmaplot computer package was used to analyze enzyme kinetics data (SigmaPlot, Systat Software, San Jose, CA, USA). To calculate the concentration of inhibitor (cantharidin) which inhibits 50% activity (IC50) of ALP, cantharidin in variable concentrations dissolved in acetone was added to the mixture containing 170 μL ALP buffer, 10 μL enzyme extract, 10 μL inhibitor and 10 μL of p-NPP solution at 37 °C. No inhibitor was added to the control. The inhibition reaction was carried at 37 °C. The IC50 value was estimated by percent inhibitory activity vs. concentration of inhibitor using Microsoft Excel, 2007.Homology modelling and molecular dynamics simulations
Template search and sequence alignment
In order to find a suitable protein crystal structure as a template, protein blast of the amino acid sequence with Helicoverpa armigera ALP (GenBank ACF40806) was carried out by Protein Blast in NCBI PDB database. After performing a BLASTP search against Protein Data Bank (PDB), several sequences homologous to our query sequence were obtained. The top hit Shrimp Alkaline Phosphatese of Pandalus borealis (SAP; PDB 1K7H)46 revealed better alignment with 43% sequence identity, 88% query coverage and E-value of 4e-132. Homology modelling to HaALP was performed using the crystal structure of SAP as a template.Ligand modelling
The three dimensional (3D) chemical structure of the cantharidin was derived from Pubchem (http://pubchem.ncbi.nlm.nih.gov). Mercury 3.0 software was used to add hydrogen atoms and coordinates were saved as MOL2 files.HaALP model preparation
Modeller was used to construct the three dimensional structure model of HaALP and results of sequence alignment was used to find the structure's conserved area using a target template, 1K7H. Modeller operation of 100 times produced 100 HaALP molecular three-dimensional models. Molecular structure with the lowest energy structure, according to DOPE (Discrete Optimized Protein Energy) score was used for further work. The structural quality of the model was assessed by Ramachandran plot in MolProbity47 which enabled us to select highest rating structure for further energy optimization and molecular dynamics simulations.HaALP molecular dynamics and energy optimization
All the molecular dynamic work was performed by NAMD 2.7b2 program. HaALP, water molecules and ions were assigned to full atomic force field CHARMM 27. During the simulations, all bond lengths containing hydrogen atoms were constrained employing the SHAKE algorithm43 and the integration time step was set to 2 fs. The HaALP ensemble was minimized by 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps with solutes constrained, followed by 20
000 steps with solutes constrained, followed by 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps minimizations without any constraint. Then, the minimized systems were slowly heated form 0 up to 310 K within 500 ps with all Cα atoms and the three zinc ions constrained. The non-constrained simulations were performed for 5 ns at a constant temperature of 310 K and a constant pressure of 1 atm through the Langevin piston method.48 Finally a well-balanced conformation was achieved for subsequent process.
000 steps minimizations without any constraint. Then, the minimized systems were slowly heated form 0 up to 310 K within 500 ps with all Cα atoms and the three zinc ions constrained. The non-constrained simulations were performed for 5 ns at a constant temperature of 310 K and a constant pressure of 1 atm through the Langevin piston method.48 Finally a well-balanced conformation was achieved for subsequent process.
Ligand-binding pocket determination
Cloning Zn1, Zn2 and Zn3 from the template SAP to HaALP was done by Modeller program to obtain the coordinates of ZN1, ZN2 and ZN3 in HaALP. The average value of atomic space coordinates of Zn1, Zn2 and Zn3 was used to get atomic space coordinate of the active centre of HaALP for docking work.Docking simulation
Molecular docking calculations were carried out using AutoDock 4.2 package. The MMFF94 force field was used for energy minimization of the ligand molecule, cantharidin before docking into the active pocket of ALP model. Gasteiger partial charges were added to the ligand atoms. Non-polar hydrogen atoms were merged and rotatable bonds were defined. Docking calculations were carried out on HaALP protein model. Essential hydrogen atoms, Kollman united atom type charges and solvation parameters were added with the aid of AutoDock tools. Affinity grid of 20 × 20 × 20 grid points and 0.375 spacing were generated using the Autogrid program (Fig. 6A). Amino acid residues of the grid are shown in Fig. 6B. AutoDock parameter and distance-dependent dielectric functions were used in the calculation of the Van der Waals and the electrostatic terms, respectively. Docking simulations were performed using the Lamarckian genetic algorithm (LGA) and the Solis & Wets local search method. Initial position, orientation and torsions of the ligand molecules were set randomly. Each docking experiment was derived from 100 different runs that were set to terminate after a maximum of 250000 energy evaluations. The population size was set to 150. During the search, a transitional step of 0.2 and quaternion and torsion steps of 5 was applied. The final solution was selected according to size of clusters and the estimated free energy of binding. The best solution of the largest cluster was selected as the best mode for each docking test. After carefully checking the docking results, the mode with the lowest value of free energy of binding was taken as the final binding mode. Ligplot program was applied to calculate the schematic representation of residue–ligand interactions between cantharidin and the active site of HaALP. | ||
| Fig. 6 The grid box of HaALP. (a) Spatial position of the active site within HaALP. (b) Letters shaded in dark black are amino acid residues in the active site of HaALP. | ||
Conclusions
Based on the collective assessment of our data we concluded that cantharidin inhibits catalytic activity of ALPs both in vivo and in vitro. The inhibitory effect of cantharidin on ALPs may be one of the reasons for its synergism with insecticides. We may also suggest its possible role in the phosphorylation of proteins with phosphoseryl and phosphothreonyl amino acid residues. In conclusion, we can suggest a role of cantharidin as insecticide synergist as one of the potential biological roles in the control of H. armigera.Acknowledgements
We sincerely appreciate Prof. J. R. Schrock (Emporia State University, USA) for revising the manuscript. This study was supported by the Special Fund for the Public Interest (Agriculture) (200903052) by The Ministry of Science and Technology, The Ministry of Agriculture, China, and the ‘13115’ Sci-Tech Innovation Project of Shaanxi Province (2007ZDKG-14).Notes and references
- M. Eguchi, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1995, 111, 151–162 CrossRef CAS.
- I. Y. Sakharov, I. E. Makarova and G. A. Ermolin, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1998, 92, 119–122 CrossRef.
- B. Horie, Bull. Sericultural Exp. Station Japan, 1998, vol. 15, pp. 275–289 Search PubMed.
- V. Mestrovic and M. Pavela-Vrancic, Biochimie, 2003, 85, 647–650 CrossRef CAS PubMed.
- D. Sarrouilhe, P. Lalegerie and M. Baudry, Biochim. Biophys. Acta, 1992, 1118, 116–122 CrossRef CAS.
- M. Kucera and J. Weiser, Acta Entomol. Bohemoslov., 1974, 71, 289–293 CAS.
- P. Sujak, K. Ziemnicki, J. Ziemnicka, J. J. Lipa and L. Obuchowicz, J. Invertebr. Pathol., 1978, 31, 4–9 CrossRef CAS.
- W. Chang, K. Zachow and D. Bentley, Development, 1993, 118, 651–663 CAS.
- M. J. Sukhanova, L. G. Grenback, N. E. Gruntenko, T. M. Khlebodarova and I. Y. Rauschenbach, J. Insect Physiol., 1996, 42, 161–165 CrossRef CAS.
- T. R. F. Wright, Adv. Genet., 1987, 24, 127–221 CAS.
- I. Y. Rauschenbach, N. A. Chentsova, A. A. Alekseev, N. E. Gruntenko, N. V. Adonyeva, E. K. Karpova, T. N. Komarova, V. G. Vasiliev and M. Bownes, Arch. Insect Biochem. Physiol., 2007, 65, 95–102 CrossRef CAS PubMed.
- I. Y. Rauschenbach, E. V. Bogomolova, N. E. Gruntenko, N. V. Adonyeva and N. A. Chentsova, J. Insect Physiol., 2007, 53, 587–591 CrossRef CAS PubMed.
- R. Srinivas, S. S. Udikeri, S. K. Jayalakshmi and K. Sreeramulu, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2004, 137, 261–269 CrossRef CAS PubMed.
- X. Gao, F. Zhang, B. Zheng, R. Wang and B. Lin, Insect Sci., 1996, 3, 243–255 CrossRef.
- R. H. Arnett, M. C. Thomas, P. E. Skelley and J. H. Frank, American Beetles, vol. 2, CRC Press, Florida, Boca Raton, USA, 2002, p. 861 Search PubMed.
- K. Dettner, G. Bauer and W. Völkl, Vertical Food Web Interactions, Springer Verlag., Berlin, Germany, 1997, p. 390 Search PubMed.
- G. D. Prestwitch and G. J. Blomquist, Pheromone Biochemistry, Academic Press, Orlando, 1987, p. 565 Search PubMed.
- Blister Beetles in Alfalfa. http://www.ca.uky.edu/entomology/entfacts/ef102.asp (15 1 2013).
- F. L. Cui, X. Li, Z. Q. Ma and Y. L. Zhang, Environ. Entomol., 2009, 31, 143–149 Search PubMed.
- Y. L. Zhang, Y. Zhou and Z. Y. Zhang, Acta Entomol. Sin., 2003, 46, 272–276 CAS.
- Y. M. Li and J. E. Casida, Proc. Natl. Sci. U. S. A., 1992, 89, 11867–11870 CrossRef CAS.
- M. J. Graziano and J. E. Casida, Toxicol. Lett., 1987, 37, 143–148 CrossRef CAS PubMed.
- N. Kawamura, Y. M. Li, J. L. Engel, W. G. Dauben and J. E. Casida, Chem. Res. Toxicol., 1990, 3, 318–324 CrossRef CAS PubMed.
- M. J. Graziano, A. L. Waterhouse and J. E. Casida, Biochem. Biophys. Res. Commun., 1987, 149, 79–85 CrossRef CAS PubMed.
- R. H. Decker, J. Invest. Dermatol., 1968, 51, 141–146 CrossRef CAS PubMed.
- F. K. Bagatell, K. Dugan and G. F. Wilgram, Toxicol. Appl. Pharmacol., 1969, 15, 249–261 CrossRef CAS PubMed.
- Y. Ma, R. R. Liu, Z. Q. Ma and Y. L. Zhang, Acta Entomol. Sin., 2010, 53, 870–875 CAS.
- S. L. Zheng, Y. L. Zhang, F. X. An and X. Zhang, Acta Phytophylacica Sin., 2007, 34, 83–86 CAS.
- Z. Bikadi, L. Demko and E. Hazai, Arch. Biochem. Biophys., 2007, 461, 225–234 CrossRef CAS PubMed.
- D. Sarrouilhe, P. Lalegerie and M. Baudry, Biochim. Biophys. Acta, 1992, 1118, 116–122 CrossRef CAS.
- P. Cohen, C. F. Holmes and Y. Tsukitani, Trends Biochem. Sci., 1990, 15, 98–102 CrossRef CAS PubMed.
- C. Bialojan and A. Takai, Biochem. J., 1988, 256, 283–290 CrossRef CAS PubMed.
- M. Suganuma, H. Fujiki, S. Okabe, S. Nishiwaki, D. Brautigan, T. S. Ingebritsen and M. R. Rosner, Toxicon, 1992, 30, 873–878 CrossRef CAS PubMed.
- J. F. Dawson and C. F. Holmes, Front. Biosci., 1999, 4, D646–D658 CrossRef CAS.
- B. Tuleva, E. Vasileva-Tonkova and D. Galabova, FEMS Microbiol. Lett., 1998, 161, 139–144 CrossRef CAS PubMed.
- U. P. Chaudhari, N. D. Trivedi, S. R. R. Patil and S. Banerjee, Int. J. ChemTech Res., 2010, 2, 122–128 CAS.
- M. Ahmed and A. R. McCaffery, Pestic. Biochem. Physiol., 1991, 41, 41–52 CrossRef.
- J. E. Carrel, J. P. Doom and J. P. McCormick, J. Chromatogr., 1985, 342, 411–415 CrossRef CAS PubMed.
- O. A. Bessey, J. Biol. Chem., 1964, 164, 321–329 Search PubMed.
- A. Sali and T. L. Blundell, J. Mol. Biol., 1993, 234, 779–815 CrossRef CAS PubMed.
- L. Kale, R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan and K. Schulten, J. Comput. Phys., 1999, 151, 283–312 CrossRef CAS.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. Van de streek and P. A. Wood, Appl. Crystallogr., 2008, 41, 466–470 CrossRef CAS.
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, J. Comput. Chem., 1998, 19, 1639–1662 CrossRef CAS.
- Z. Bikadi and E. Hazai, J. Cheminf., 2009, 11, 15 Search PubMed.
- A. C. Wallace, R. A. Laskowski and J. M. Thornton, Protein Eng., 1995, 8, 127–134 CrossRef CAS PubMed.
- M. E. De Backer, S. McSweeney, H. B. Rasmussen, B. W. Riise, P. Lindley and E. Hough, J. Mol. Biol., 2002, 318, 1265–1274 CrossRef CAS PubMed.
- V. B. Chen, W. B. Arendall 3rd, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson and D. C. Richardson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 12–21 CrossRef CAS PubMed.
- W. D. Cornell, P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell and P. A. Kollman, J. Am. Chem. Soc., 1995, 117, 5179–5197 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |