Chemical computing based on Turing patterns in two coupled cells with equal transport coefficients
Abstract
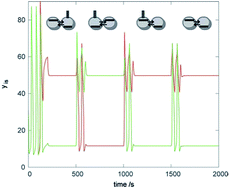
We study instabilities leading to inhomogeneous stationary states (discrete Turing patterns) using a simple model of glycolytic oscillatory reaction in two mass-coupled stirred open reactors with equal coupling coefficients of the activator and inhibitor and explore their ability to serve as logical devices. The system represents the simplest coupled cell system. It is found to possess coexisting stable homogeneous oscillations and a pair of stable complementary asymmetric Turing patterns. We examine transitions from the oscillatory regime to Turing patterns, or from one pattern to another, elicited by targeted perturbations specific for each type of transition. We further develop our earlier proposition of using the Turing patterns in chemical computing and explore logical functions available in the two cell system. Digital memory storage, a sensor, a signal conducting module and simple logic gates using NAND, AND, OR, NOR, and NOT functions are found. The main advantage of the two-cell system is that it can serve as a basic unit in the construction of a lattice representing a universal chemical processor, whereas larger linear or circular arrays are more suitable for tailor-made operations.

 Please wait while we load your content...
Please wait while we load your content...