Simultaneous co-ordination of three cytosine ligands displaying different binding sites around the copper centres†
Yogesh Prakash Patil and
Munirathinam Nethaji*
Inorganic and Physical Chemistry Department, Indian Institute of Science, Bangalore 560012, India. E-mail: mnetaji@ipc.iisc.ernet.in
First published on 28th August 2014
Abstract
We report the synthesis and structural characterization of a polymeric ternary copper–cytosine–phenanthroline complex, [Cu4(phen)3(μ3-cyt)2(μ-OH)(cyt)(OH)Cl3]n·16H2O, where three cytosine ligands with different binding sites have simultaneously complexed to the four copper metal centres. Interestingly, the complex exhibits two different coordination geometries around the metal centres.
Interactions of metal ions with nucleic acid constituents has been a topic of interest for the past few decades.1,2 The major interest arises as metal–DNA interactions play a crucial role in DNA stabilisation [bivalent metals like Zn(II), Mg(II)],3 destabilisation [Cu(II), Co(II), Cd(II)],3 mutagenesis and also in variety of enzymatic processes.4,5 Nucleobases, a part of nucleic acids, are multidentate ligands with different binding sites which vary with metal ions and physiological conditions. Thus it is of immense interest to document the underlying principles which dictate the propensity of the binding sites of nucleobases to metals.
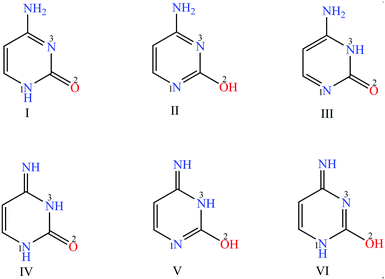
To understand these interactions several crystallographic studies are done over the past few decades with different metals and nucleobases. For instance the nucleobase cytosine (tautomer I, Fig. 1), interacts with alkali metals like Na+ and Ca2+ by coordinating through the exocyclic oxygen atom O2 displaying the oxyphilic nature.6,7 Whereas, the transition metals like Rh3+, Cd2+, Co2+, Zn2+, Ni2+, Pt2+ coordinate to cytosine both in binary and ternary complexes through the oxygen and/or nitrogen binding sites.8–20
Cytosine is also known to interact with copper(II) metal ion thereby breaking hydrogen bonds in between the DNA base pair and destabilizing its structure.21 In contrast, DNA base pairing via copper(II) coordination to the artificial bases like pyridine-2,6-dicarboxylate and pyridine forms a stable and selective helical structure.22 These features could be well understood with a detailed analysis of copper cytosine complexes. The structural studies done so far with copper cytosine binary21,23,24 and ternary25,26,29 complexes reveal monodentate cytosine (tautomer I, Fig. 1) coordination through nitrogen atom, N3 of the base with a square planar geometry around the copper centre. Till date, to our knowledge, there have been no structure reports for any other binding modes observed in case of copper cytosine binary/ternary complexes. In this context the structure elucidation of binary/ternary copper–cytosine complexes is pivotal to shed light on the other plausible binding modes of cytosine with copper centre/s. Herein, we present synthesis and structure elucidation of ternary polymeric copper complex, [Cu4(phen)3(μ3-cyt)2(μ-OH)(cyt)(OH)Cl3]n·16H2O (1a), with cytosine and 1,10-phenanthroline as coligands. This complex was originally designed as a part of library of compounds targeted as models for protein–metal–nucleic acid interactions, where 1,10-phenanthroline act as side chain mimic of proteins.27,28
Complex 1a was prepared by mixing aqueous solution of Cu2(phen)2(μ-Cl)2Cl2 (1) (50 mg, 0.08 mmol) with 5 ml aqueous solution of cytosine (26.5 mg, 0.24 mmol) and stirring the mixture for 17 hours [see ESI†]. The plate shaped skyblue colored crystals of 1a were obtained by slow evaporation of the reaction mixture after ∼2 weeks. Importantly, the yield of the complex was very low (11.5%).
Complex 1a crystallizes in the orthorhombic space group Pbca with Z = 8. The ORTEP diagram is shown in Fig. 2 while, the selected bond length and angles around the copper centres are summarised in ESI (Table S1†).
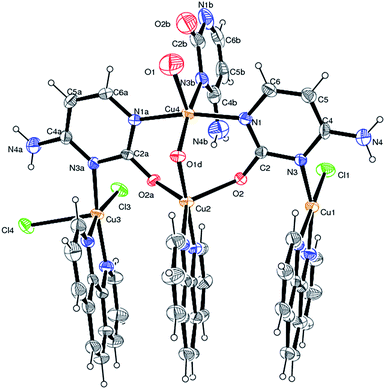 | ||
| Fig. 2 ORTEP diagram of 1a with 20% probability for the sake of clarity.‡ | ||
The crystal structure of 1a consists of a polymeric copper(II) complex with cytosine and 1,10-phenanthroline as co-ligands (Fig. S1;† ESI). The complex is neutral with the charge balanced by the coordinating ligands; which are three chloride ions, two hydroxyl ions and three uninegative cytosine bases. Among the three cytosine rings two coordinate in a tridentate fashion while the third cytosine ring depicts monodentate coordination.
This observation poses two questions (1) are the three cytosines are same or different and (2) which of the possible cytosine tautomer/s (Fig. 1) have coordinated to the copper centers. This can be resolved by a meticulous comparison of the geometry of cytosine rings with each other and with the free cytosine tautomeric forms I31 and III32 (major and minor tautomers observed in solid and solution state) [Table S2;† ESI]. This approach has been previously used by Lippert and coworkers to confirm the presence of minor tautomer III in case of the platinum cytosine complexes.14 The analysis of the cytosine rings in 1a suggests that all the rings are same and they concur with the deprotonated form of tautomer I. The details of the analysis are described as follows.
The three independent cytosine rings in 1a are labeled as A, B and C for the ease of discussion (Fig. 3). The comparison of geometries for the three cytosine rings show that the differences in bond lengths do not exceed above the 3σ level.33 Similarly all the bond angles but for one are comparable (differences <3σ). The external angle ∠N3–C2–O2 is slightly larger in cytosine ring C as compared to that in A and B (max. 5.4σ). This may be due to the involvement of A and B in the tridentate coordination to copper centers. The above calculation shows that all the three cytosine rings are same. Further each cytosine ring was compared with the geometries of anhydrous cytosine [tautomer I] and the results are as follows. (1) Most of the bond angles of A, B and C are consistent with the tautomeric form I (differences <3σ). Especially, there is an agreement for all the angles of C with tautomer I except for ∠C5–C6–N1 which is slightly larger in C (4.1σ). This means that the ring geometry for C (consistent with tautomer I) is not distorted upon coordination to the copper centre. There are deviations in three angles for A and B which may be a result of the involvement of A and B in tridentate coordination.34 and (2) all the bond lengths for A, B and C are comparable with tautomer I except for few in A and B.34 The geometries for A, B and C were also compared with the minor tautomer III (calculated for gas phase14) and it reveals that there are significant deviations for both bond lengths (deviation upto 0.10 Å) and bond angles (deviation upto 8.4°). Thus, to conclude all the three cytosine rings corroborate with the tautomeric form I with the deprotonation occurring at N1 nitrogen atom.
 | ||
| Fig. 3 Two different geometries around copper centres as seen from the tau value [τ = (α − β)/60 where α and β are angles between the trans ligand of the of square pyramidal base geometry].30 | ||
The monomeric unit of complex 1a (shown in Fig. 2 and 3) can be described as follows. Two copper centres Cu1 and Cu3 have same coordination environment viz the basal plane of the square pyramid is formed by cytosine [N3 and N3A], 1,10-phenanthroline [N7, N8 and N11, N12] and chloride ligands [Cl1, Cl3] while the axial site is occupied by other chloride ion [Cl4] which acts as a bridge between Cu1 and Cu3 in the polymeric chain. The Cu–N distances of three cytosine rings are approximately 2.02 Å which concur with the reported Cu–N distance for copper cytosine complexes.25,26,29 The two cytosine rings (conjugate base of tautomer I) attached to Cu1 and Cu3 act as tridentate ligand coordinating to two other copper centres [Cu2, Cu4] via O2, O2A and N1, N1A respectively. The remaining three sites of Cu2 are occupied by 1,10-phenanthroline [N9, N10] and a bridged hydroxyl [O1D] moiety coordinating to Cu2 and Cu4 simultaneously. The third cytosine (conjugate base of tautomer I) is coordinated to Cu4 via N3 nitrogen atom acting as monodentate ligand.
Complex 1a exhibits some interesting features. There are four copper atoms in the monomeric unit of the polymer 1a, which exhibits two type of coordination geometries about the copper centres. Out of the four, three copper centres adopt square pyramidal [4 + 1] geometry [τ = 0.17 (Cu1), 0.028 (Cu3) and 0.053 (Cu4)], whereas the fourth copper centre exhibits distorted trigonal bipyramidal [3 + 2] geometry30 [τ = 0.66 (Cu2)] (Fig. 3). It is noteworthy that three copper atoms Cu1, Cu2 and Cu3 which are connected by the two cytosine rings are having the 1,10-phenanthroline rings coordinated on the other side whereas Cu4 does not have this environment.
The analysis of least square planes shows that the copper atom Cu1 deviates from the distorted basal plane [Cu1 = 0.104(1) Å], while Cu2, Cu3 and Cu4 deviate from the basal plane [Cu2 = 0.146(1) Å, Cu3 = −0.195(1) Å, Cu4 = 0.219(1) Å] (Table S3;† ESI). The unit cell packing shows that the molecules are held by both intra and intermolecular non-covalent interactions (Table S4;† ESI). The structure is stabilized by slipped π–π intramolecular stacking interactions between the phenanthroline rings (Table S5;† ESI). The cytosine moieties interact with each other through bifurcated N–H⋯O hydrogen bond where the proton of N6b is involved with O2 and O2A of the other two cytosine moieties coordinated to the same copper centre. The monomeric unit also shows the presence of N–H⋯Cl hydrogen bonds between bridged cytosine and axial chloride ion (Fig. S2;† ESI). The neighboring chains of the polymer 1a are linked by intermolecular slipped π–π stacking interactions between the cytosine ring attached to Cu4 and the phenanthroline rings. The chains are also connected through C–H⋯Cl hydrogen bonds where the chlorine atom Cl4 is involved in the bifurcated hydrogen bond, one as intramolecular and the second as intermolecular interaction (Fig. S3;† ESI). Both the Nitrogen atoms [N4, N4A] of different cytosine rings are involved in the noncovalent interactions, with the water [O41, O10W] as intermolecular and with chlorine atoms Cl4 and Cl4* (* symmetry generated) as intramolecular hydrogen bond respectively. The water molecules pack between the polymeric chains via noncovalent interactions (Fig. S4;† ESI). These water molecules do not interact with the polymeric strands and fill the void space in between the strands. The polymer chain which runs along the c axis displays hydrophobic and hydrophilic regions. The dangling cytosine moieties are facing along the −a axis while the phenanthroline rings are facing the opposite side +a axis. The water molecules are sandwiched between the chains surrounding the cytosine moiety. Thus the channels of hydrogen bonded water molecules interconnect the cytosine rings. Notably, these water molecules are trapped between the polymers make extensive network via possible hydrogen bonds (hydrogen atoms attached to the water molecules could not be located from the difference Fourier map) (Table S6;† ESI).
Complex 1a contains a large number of water molecules as solvent of crystallization (16 molecules per asymmetric unit). So, when the crystal was heated gradually at 40 K per hour in situ and the diffraction data at different temperature was recorded, changes in the unit cell dimensions were observed (Table S7,† ESI). It is seen that the cell volume decreases initially at 308 K as compared to room temperature data (293 K). This can be attributed to loss of loosely bound water molecules as described earlier. Further heating leads to change in the crystal system from orthorhombic (308 K)–tetragonal (318 K)–orthorhombic C (328 K)–tetragonal (338 K) to tetragonal (348 K) system.
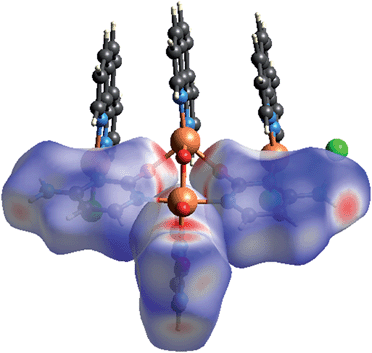
To quantitatively ascertain these interactions Hirshfeld surface analysis of the three cytosine rings was done.36,37 The dnorm surface for the three cytosine rings were obtained independently for the monomeric unit of polymer 1a and fingerprint break down values of these rings were compared (Fig. 4). It may be noted that fingerprint analysis, though generally used to quantify the proportions of different intermolecular interactions, in the present case has been used to quantify the differences in metal–ligand binding modes. The area of N⋯Cu coordination regions in A and B cytosine rings are twice the corresponding area in C. This is due to the tridentate coordination of A and B with the copper centres (Table S8;† ESI). The C⋯C interactions which correspond to π–π stacking interaction is 8.0% for cytosine ring C as it is involved in π–π stacking with neighboring polymeric chains while it is absent for A and B. The contribution of H⋯O interactions which can be either C–H⋯O, O–H⋯O and N–H⋯O vary for the three cytosine rings. Cytosine C has about 8.9% contribution of O⋯O interaction which can be attributed to O–H⋯O interaction with the solvent water molecule and coordinated hydroxyl ion concurring with the hydrophilic nature of the surroundings near cytosine C as mentioned earlier.
A possible mechanism of the formation of the polymer 1a can be conceived as involving the following steps: (i) the precursor 1 can react in presence of cytosine (tautomer I) to give V and can also form species IV via II and III (Fig. 5), and (ii) further the species IV and V react with cytosine (tautomer I) to form the polymer 1a. It should be noted that the species IV and V mentioned were not detected experimentally; the crystals for II and III were obtained as a cocrystal (reported earlier35) in the same reaction mixture of complex 1a, supporting this hypothesis. The cell dimensions for the cocrystal are a = 8.789(3) Å, b = 9.370(1) Å, c = 16.488(5) Å, α = 105.48(1)°, β = 99.12(1)°, γ = 101.51(2)°.
In summary, complex 1a presents a unique example where the copper centres have two different coordination geometries and also cytosine rings depict different modes of coordination. In all the earlier crystal structure reports on copper–cytosine binary and ternary complexes, cytosine was known to coordinate via N3 nitrogen atom only. Herein we demonstrate, for the first time, that with copper metal other binding sites (N1, N3 and O2) for cytosine are also possible. The formation of complex 1a also points to the fact that two different binding modes (monodentate and tridentate) of cytosine can be present together. This finding enriches the existing knowledge of metal nucleobase interaction. In addition this is a first example of a homometallic coordination polymer with copper and nucleobase cytosine.
Acknowledgements
Yogesh P. Patil would like to thank CSIR, New Delhi, India for the award of Senior Research Fellowship.Notes and references
- P. D. Meester, D. M. L. Goodgame, K. A. Price and A. C. Skapski, Nature, 1971, 229, 191–192 CrossRef.
- J. Barton, Science, 1986, 233, 727–734 CrossRef CAS.
- A. Višnjevac, N. Biliškov and B. Žinić, Polyhedron, 2009, 28, 3101–3109 CrossRef PubMed.
- J. Müller, R. K. O. Sigel and B. Lippert, J. Inorg. Biochem., 2000, 79, 261–265 CrossRef.
- M. A. Salam and K. Aoki, Inorg. Chim. Acta, 2000, 311, 15–24 CrossRef CAS.
- D. Armentano, G. De Munno and R. Rossi, New J. Chem., 2006, 30, 13–17 RSC.
- K. Ogawa, M. Kumihashi, K. Tomita and S. Shirotake, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1980, 36, 1793–1797 CrossRef.
- K. Aoki and M. A. Salam, Inorg. Chim. Acta, 2001, 316, 50–58 CrossRef CAS.
- M. C. Capllonch, A. García-Raso, A. Terrón, M. C. Apella, E. Espinosa and E. Molins, J. Inorg. Biochem., 2001, 85, 173–178 CrossRef CAS.
- G. De Munno, M. Medaglia, D. Armentano, J. Anastassopoulou and T. Theophanides, J. Chem. Soc., Dalton Trans., 2000, 1625–1629 RSC.
- D. Badura and H. Vahrenkamp, Inorg. Chem., 2002, 41, 6013–6019 CrossRef CAS PubMed.
- G. Cervantes, J. J. Fiol, A. Terron, V. Moreno, J. R. Alabart, M. Aguilo, M. Gomez and X. Solans, Inorg. Chem., 1990, 29, 5168–5173 CrossRef CAS.
- S. Jaworski, H. Schöllhorn, P. Eisenmann, U. Thewalt and B. Lippert, Inorg. Chim. Acta, 1988, 153, 31–38 CrossRef CAS.
- W. Brüning, E. Freisinger, M. Sabat, R. K. O. Sigel and B. Lippert, Chem.–Eur. J., 2002, 8, 4681–4692 CrossRef.
- W. Brüning, I. Ascaso, E. Freisinger, M. Sabat and B. Lippert, Inorg. Chim. Acta, 2002, 339, 400–410 CrossRef.
- W. Brüning, R. K. O. Sigel, E. Freisinger and B. Lippert, Angew. Chem., Int. Ed., 2001, 40, 3397–3399 CrossRef.
- E. Gil Bardají, E. Freisinger, B. Costisella, C. A. Schalley, W. Brüning, M. Sabat and B. Lippert, Chem.–Eur. J., 2007, 13, 6019–6039 CrossRef PubMed.
- A. Khutia, P. J. Sanz Miguel and B. Lippert, Inorg. Chem., 2010, 49, 7635–7637 CrossRef CAS PubMed.
- A. Khutia, P. J. Sanz Miguel and B. Lippert, Chem.–Eur. J., 2011, 17, 4195–4204 CrossRef CAS PubMed.
- A. Khutia, P. J. Sanz Miguel and B. Lippert, Chem.–Eur. J., 2011, 17, 4205–4216 CrossRef CAS PubMed.
- A. Panfil, A. Terron, J. J. Fiol and M. Quiros, Polyhedron, 1994, 13, 2513–2518 CrossRef CAS.
- S. Atwell, E. Meggers, G. Spraggon and P. G. Schultz, J. Am. Chem. Soc., 2001, 123, 12364–12367 CrossRef CAS PubMed.
- D. Tran Qui and E. Palacios, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1990, 46, 1220–1223 CrossRef.
- M. Palaniandavar, I. Somasundaram, M. Lakshminarayanan and H. Manohar, J. Chem. Soc., Dalton Trans., 1996, 1333–1340 RSC.
- A. García-Raso, J. J. Fiol, A. López-Zafra, A. Tasada, I. Mata, E. Espinosa and E. Molins, Polyhedron, 2006, 25, 2295–2302 CrossRef PubMed.
- B. García, J. Garcia-Tojal, R. Ruiz, R. Gil-García, S. Ibeas, B. Donnadieu and J. M. Leal, J. Inorg. Biochem., 2008, 102, 1892–1900 CrossRef PubMed.
- I. Samasundaram, M. K. Kommiya and M. Palaniandavar, J. Chem. Soc., Dalton Trans., 1991, 2083–2089 RSC.
- A. Terrón, J. J. Fiol, A. García-Raso, M. Barceló-Oliver and V. Moreno, Coord. Chem. Rev., 2007, 251, 1973–1986 CrossRef PubMed.
- T. J. Kistenmacher, T. Sorrell and L. G. Marzilli, Inorg. Chem., 1975, 14, 2479–2485 CrossRef CAS.
- A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn and G. C. Verschoor, J. Chem. Soc., Dalton Trans., 1984, 1349–1356 RSC.
- R. J. McClure and B. M. Craven, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1973, 29, 1234–1238 CrossRef CAS.
- C. Colominas, F. J. Luque and M. Orozco, J. Am. Chem. Soc., 1996, 118, 6811–6821 CrossRef CAS.
- The ESD is calculated according to σ = (σ12 + σ22)1/2 with σ1 and σ2 being the errors in bond lengths and angles which are compared.
- ∠C6–N1–C2, ∠N3–C2–O2 shorter than tautomer I (5.2–8.7σ); ∠N1–C2–N3 larger than tautomer I (6.5–7σ); C2–N1 in A shorter than tautomer I (4.9σ); C4–N3 in A longer than tautomer I (3.9σ); C2–O2 of A and B larger than tautomer I (3.7–4.6σ).
- Li-P. Lu, M.-L. Zhu and J. P. Yang, J. Inorg. Biochem., 2003, 95, 31–36 CrossRef CAS.
- J. J. McKinnon, D. Jayatilaka and M. A. Spackman, Chem. Commun., 2007, 3814–3816 RSC.
- M. A. Spackman and D. Jayatilaka, CrystEngComm, 2009, 11, 19–32 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic methods, variable temperature experiment, figures and tables. CCDC 951698. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4ra07828d |
‡ Crystal data for 1a: C48H70Cl3Cu4N15O21, Mr = 1553.7, sky blue plate, 0.41 × 0.04 × 0.01 mm3, orthorhombic, Pbca, a = 20.653(5) Å, b = 24.066(5) Å, c = 26.128(5) Å, V = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 987(5) Å3, Z = 8, ρc = 1.589 g cm−3, μ = 1.499 mm−1, R1 = 0.0683 (F2 > 2s), wR2 = 0.1888 (all data), 11 987(5) Å3, Z = 8, ρc = 1.589 g cm−3, μ = 1.499 mm−1, R1 = 0.0683 (F2 > 2s), wR2 = 0.1888 (all data), 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 independent reflections (6312 with F2 > 2s) and 815 parameters. 438 independent reflections (6312 with F2 > 2s) and 815 parameters. |
| This journal is © The Royal Society of Chemistry 2014 |