DOI:
10.1039/C4RA07788A
(Paper)
RSC Adv., 2014,
4, 48399-48410
Preparation of quantum dot-embedded polymeric nanoparticles using flash nanoprecipitation
Received
29th July 2014
, Accepted 23rd September 2014
First published on 24th September 2014
Abstract
We developed a unique and efficient method to encapsulate quantum dots within amphiphilic polymer micelles using the flash nanoprecipitation technique and various micromixers (multi-inlets vortex mixer, MIVM, and confined impinging-jet mixer, CIJM). Owing to the rapid homogeneous mixing provided by the micromixers, we produced highly stable QD-embedded particles with very narrow size distributions. By adjusting the solvent-to-nonsolvent ratio, homopolymer concentration, and micromixer arrangement, the resulting mean particle diameter was varied from 90 to 800 nm. Additionally, the self-quenching of quantum dots, which is commonly observed in confined systems, was nearly eliminated by adding hydrophobic homopolymer during mixing. This technique results in controllable, rapid synthesis of fluorescent polymeric particles that are stable for many months. Potential applications of this method include the efficient synthesis of fluorescent tracer particles, tags for optical barcoding applications, and biosensors that utilize fluorescence resonance energy transfer (FRET).
1 Introduction
Luminescent quantum dots (QDs) have been used widely in many biological applications including cellular targeting and imaging,1 in vivo animal imaging,2 biosensors,3,4 and multicolor microarrays5 due to their unique optical properties. Compared to conventional fluorophores such as organic dye molecules, QDs have broad absorption, narrow size-tunable and symmetric emission spectra, and large absorption cross-sections. Their size-dependent optical properties are attributed to the three-dimensional quantum confinement of charge carriers (electrons and holes). These superior properties endow them unique photophysical characteristics having considerable advantages over conventional dyes in many applications including an unusually high resistance to photobleaching.
Encapsulating QDs into microspheres or nanobeads is very attractive for a wide range of applications including light–matter interactions,6 semiconductor microlasing,7 and biological markers.8 In optical barcoding, which requires quantitatively embedding fluorophores with different colors into individual beads, the broad absorption of QDs is a significant advantage because only a single excitation source is required to induce fluorescence among all fluorophores having distinct emission colors. In addition, the narrow emission spectra of QDs increases the number of attainable barcode combinations.9
The recent literature contains myriad methods for internalizing QDs into beads. The most popular current approaches include the following general categories: (1) embedding hydrophobic QDs through emulsion and dispersion polymerization of styrene10 or acrylates;11 (2) coupling QDs to polymer microspheres via polymerizable surface ligands (phosphine-based12,13); (3) incorporation of QDs by swelling of polymer beads in a solvent–nonsolvent solution;9 (4) entrapment of QDs via volumetric phase transitions of polymer gels;14 (5) immobilization of charged QDs inside polymer microspheres via electrostatic interactions;15,16 (6) encapsulation of QDs into polymers using a layer-by-layer deposition technique,17 and (7) embedding QDs into block copolymer micelles18–21 or silica beads.22–24 However, these methods commonly suffer from low loading capacity, broad size distributions, and/or fluorescence quenching (∼75% luminescence loss) due to the aggregation of QDs or surface modification.22 Although Yang and co-workers25 reported that they were able to achieve high loading and narrow size dispersity simultaneously, the quantum yield loss during the process was still relatively large (∼25% decrease). Moreover, the preceding processes to make these QD embedded polymer spheres are often time-consuming and costly which is perhaps the most important practical factor.
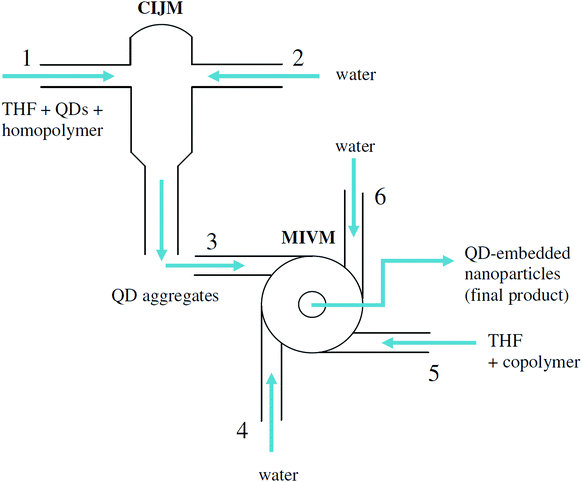
With these limitations in mind, we explored a new method based on the recent particle synthesis technique of flash nanoprecipitation (as shown in Fig. 1), which was first reported by Johnson and Prud'homme.26 In flash nanoprecipitation, hydrophobic QDs and amphiphilic polymer are dissolved into a suitable organic solvent (tetrahydrofuran, THF) that is miscible with water (nonsolvent). The rapid and turbulent mixing of solvent and nonsolvent induces the spontaneous precipitation of QD-internalized polymer micelles having tunable size and narrow size distributions depending on precise mixing conditions. Flash nanoprecipitation requires that the mixing time (τmix) is much shorter than the aggregation time (τagg) for polymer micelles (as shown in Fig. 1). The microreactors used in this study (multi-inlet vortex mixer, MIVM,27 and confined impinging jets mixer, CIJM,28 as shown in Fig. 2), which have very short mixing times (in the millisecond range), both meet this requirement. We used these microreactors to obtain a wide range of sizes (90 to 800 nm) of QD-embedded nanoparticles by adjusting the solvent-to-nonsolvent ratio and adding homopolymer during mixing. This microreactor-based approach is fast (the process can be accomplished within a few minutes) and flexible, and only hydrophobic QDs (requiring no additional pre-processing steps) are needed. Theoretically, many types of copolymers with an appropriate hydrophobic-hydrophilic balance are compatible with the technique. We further found that the self-quenching of QDs was dramatically reduced along with size polydispersity through the addition of hydrophobic homopolymers. Our goal of this work is to explore methods for synthesizing nanoparticles in a wide range, especially towards the larger size, which can broaden QDs' use in other applications such as Particle Image Velocimetry.
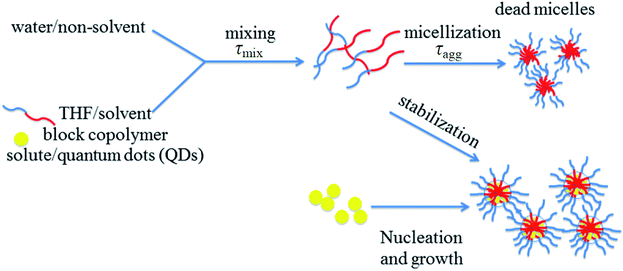 |
| | Fig. 1 Schematic of flash nano-precipitation.26 | |
 |
| | Fig. 2 Design of multi-inlet vortex mixer (MIVM) (left) and confined impinging-jet mixer (CIJM) (right). | |
2 Experimental methods
2.1 Materials
Hexadecylamine (HDA, 90%), hexamethyldisilathiane (TMS2S), trioctyl phosphine (TOP, 90%), diethylzinc (Zn 52.0 wt%) were purchased from Sigma-Aldrich (St. Louis, MO) and used as received. Cadmium acetylacetonate (Cd(acac)2) and selenium shot (Se, 99.99%) were used as received from Strem Chemicals. Trioctyl phosphine oxide (TOPO, 98%) was obtained from Alfa Aesar and used as received. Poly(styrene-b-ethylene oxide) diblock copolymer (PS-b-PEG, 9.5 kDa-b-5 kDa, Mw/Mn = 1.05) was purchased from Polymer Source (Montreal, Quebec, Canada). Polystyrene homopolymers (Mn = 5 kDa, 13 kDa, and 26 kDa) were a kind gift from the Macosko group at the University of Minnesota. Poly(diethylaminoethylmethacrylate) (PDEAEM)/Pluronic F127 pentablock copolymers and Pluronic F127 were provided by the Mallapragada group at Iowa State University. Polystyrene homopolymers (Mw = 98 kDa) were supplied by the Cochran group at Iowa State University.
2.2 CdSe–ZnS QD synthesis
The quantum dots (QDs) used in this study, synthesized using the approach reported by Clapp et al.29 and Zhang et al.,30 included CdSe core-only and CdSe–ZnS core–shell nanocrystals. Briefly, during core synthesis, Cd precursor (Cd(acac)2) and Se precursor (1 M TOP:Se) are quickly injected into a three-neck flask which contains TOP, TOPO, and HDA at high temperature (∼350 °C) under dry N2 atmosphere. The precursors quickly decompose and spontaneously nucleate CdSe nanocrystals which continue to grow until the temperature is appropriately lowered (<150 °C). After annealing overnight (∼80 °C), the cores are centrifuged to remove excess unreacted precursors. To protect the core from oxidation and passivate the surface, CdSe cores can be overcoated with multiple ZnS layers (typically ∼4–5 layers) using diethylzinc and TMS2S as precursors. The calculated amount (depending on the size of the core and the number of layers desired) of zinc and sulfur precursors are slowly added (0.4 mL min−1) to the flask containing TOPO and CdSe cores at relatively low temperature (∼150 °C) through a gas tight syringe where the flow rate is accurately controlled by a computer controlled syringe pump. Once the addition is complete, the core–shell QDs are cooled to ∼80 °C and again allowed to anneal overnight under an N2 atmosphere.
2.3 Encapsulation Method I (particles <300 nm in diameter)
The first method produces the smallest particles and largely follows the original mixing protocol for the multi-inlet vortex mixer (MIVM) described in previous reports.27 Methanol (anti-solvent) was added to QDs stored in their growth solution to precipitate the nanocrystals and remove excess TOP and TOPO ligands. This was followed by centrifugation where the supernatant was discarded and the remaining precipitated QDs were dried with N2. Dry QD powders were weighed and dissolved into THF to a final concentration of 10 mg mL−1. The specific feed arrangement of the four inlets of the MIVM is shown in Fig. 3. For the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 solvent-to-nonsolvent ratio (S
9 solvent-to-nonsolvent ratio (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS) case, two streams (streams 1 and 3) delivered a total of 45 mL of water (nonsolvent, NS), stream 2 flowed 5 mL of THF (solvent, S) with 20 mg of dissolved QDs, and stream 4 flowed 5 mL of THF with 30 mg of dissolved block copolymer. The ratio of solvent to nonsolvent was 1
NS) case, two streams (streams 1 and 3) delivered a total of 45 mL of water (nonsolvent, NS), stream 2 flowed 5 mL of THF (solvent, S) with 20 mg of dissolved QDs, and stream 4 flowed 5 mL of THF with 30 mg of dissolved block copolymer. The ratio of solvent to nonsolvent was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (S
9 (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1
NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) to ensure supersaturation conditions and rapid nanoprecipitation. The flow rates of streams 1 and 3 were 70 mL min−1, while that of streams 2 and 4 were 7.8 mL min−1. In the 1
9) to ensure supersaturation conditions and rapid nanoprecipitation. The flow rates of streams 1 and 3 were 70 mL min−1, while that of streams 2 and 4 were 7.8 mL min−1. In the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 S
1 S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS case, the delivered volumes were matched at 5 mL in all four streams. The flow rate of each stream was controlled by a syringe pump. All four streams were set to start and stop simultaneously. The product stream leaving the MIVM was collected in a vial where samples were later characterized by a Malvern Zetasizer Nano-ZS90.
NS case, the delivered volumes were matched at 5 mL in all four streams. The flow rate of each stream was controlled by a syringe pump. All four streams were set to start and stop simultaneously. The product stream leaving the MIVM was collected in a vial where samples were later characterized by a Malvern Zetasizer Nano-ZS90.
 |
| | Fig. 3 Encapsulation Method I using MIVM (<300 nm mean particle diameter). | |
2.4 Encapsulation Method II (particles 300–500 nm in diameter)
The scheme of Encapsulation Method II is shown in Fig. 4. This protocol employs the CIJM and MIVM in series such that QDs form self-aggregates in the CIJM followed by encapsulation by polymer in the MIVM. The protocol was found to produce a larger average diameter than Method I. The outlet of the CIJM was connected to one of the inlets of the MIVM with a very short section of tubing (approximately 1 inch in length). Stream 1 delivered 5 mL of THF with 20 mg QDs and 8 mg of dissolved homopolymer. Stream 2 delivered 5 mL of water. Stream 3 (outlet) delivered resulting QD aggregates that formed by mixing in the CIJM; this stream was then delivered to an inlet port of the MIVM via the short tubing. Two symmetric streams which contained 5 mL of water each (streams 4 and 6) were mixed with stream 3 containing 5 mL of THF with 30 mg of dissolved diblock copolymer (stream 5) in the central mixing chamber of the MIVM. The flow rates of all four inlet streams were identical (22.4 mL min−1). The QD-embedded nanoparticles formed in the MIVM were collected in a vial containing 90 mL of ultrapure water to suppress post-mixing Ostwald ripening.
 |
| | Fig. 4 Encapsulation Method II using CIJM (first mixer) and MIVM (second mixer) (300–500 nm mean particle diameter). | |
2.5 Encapsulation Method III (500–800 nm)
The procedure of Encapsulation Method III is conceptually similar to Method II where QDs are allowed to form self-aggregates in the CIJM. However, rather than connecting the two reactors with short tubing, the product stream from the CIJM (stream 3 in Fig. 5) was first collected in a vial, allowed to settle for a prescribed amount of time (as determined from preliminary calibration tests), and later pumped into the second mixer as an inlet stream where QD aggregates were overcoated and stabilized within block copolymer. The composition of all six streams using this method was identical to Method II.
 |
| | Fig. 5 Encapsulation Method III using CIJM (first mixer) and MIVM (second mixer) (500–800 nm mean particle diameter). | |
2.6 Size and size distribution measurement
The mean size and size distribution of these QD-embedded nanoparticles produced by the micromixers were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano-ZS90. The intensity correlation curve was collected at 25 °C at a scattering angle of 90°. The Z-average sizes and polydispersity indices (PDI) were determined by fitting the autocorrelation curve to the following expressions:31–35| |
 | (1) |
| |
 | (2) |
| |
 | (3) |
where A is the amplitude, B is the baseline value at infinite time, q is the known scattering vector, τ is the delay time, D the particle diffusion coefficient, k the Boltzmann constant, T the absolute temperature, η the viscosity of the medium, and RH the hydrodynamic radius. The hydrodynamic size extracted using this method is an average value, weighted by the particle scattering intensity. The cumulant size is defined as the Z-average size. The polydispersity index (PDI) from the cumulants algorithm is representative of the width of the hypothetical monomodal distribution, so a large PDI can indicate either a wide distribution or a multi-modal distribution. For PDI over 0.5, i.e., multimodal samples, Z-average sizes are no longer considered useful for describing general particle size characteristics.
2.7 pH stability of QD-embedded nanoparticles (Pluronic F127)
QD-embedded nanoparticles used in pH stability experiments were generated using Encapsulation Method I where QDs were embedded in Pluronic F127 copolymer. The fluorescence intensity of QD-embedded nanoparticles in varying pH buffers was measured by a Fluoromax-4 dual monochromator spectrofluorimeter (Horiba Jobin-Yvon). Buffers of varying pH were prepared by adjusting the ratio of 0.2 M sodium phosphate to 0.1 M citric acid.
3 Results and discussion
3.1 Parametric effects on particle size
Parameters that can affect the particle size and PDI include reactor configuration, flow rate of each stream, the ratio of solvent to nonsolvent, type of block copolymer and the presence of homopolymer.
As Fig. 2 (top) shows, the MIVM has four inlets and one outlet from the center of the mixing chamber. The Reynolds number (Re) for this mixer is defined in the mixing chamber as:28
| |
 | (4) |
where
ui is the velocity of
ith inlet stream,
d is the diameter of mixing chamber,
νi is the kinematic viscosity of
ith inlet stream, and
N is the total number of inlet streams (
N = 4 in this mixer). The inlets are oriented tangential to the central mixing chamber so that the momentum of each inlet stream can be varied independently. The effective Reynolds number is defined as a linear combination of the individual stream conditions. The particle size is found to be independent of flow rate once Re is above 1600 in this particular reactor suggesting that Re > 1600 is sufficient for homogeneous mixing. A low Re will produce larger particles but a broader PDI.
36
Fig. 2 (bottom) shows the design of the CIJM, which has two inlets and one outlet. The Re is defined in one inlet stream as:37
| |
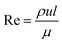 | (5) |
where
ρ is the density of the mixed stream,
u is the inlet velocity,
l is the hydraulic diameter, and
μ is the viscosity of the stream. Compared to the design of the MIVM, the CIJM requires equal momenta in both inlet streams. A Reynolds number above 500 ensures homogeneous mixing.
37,38 These two reactors produce characteristic mixing times on the order of milliseconds to achieve flash nanoprecipitation.
The general scheme of flash nanoprecipitation is shown in Fig. 1. An inlet stream containing organic solvent (THF) with dissolved hydrophobic QDs and block copolymers collides with a second stream, consisting of ultrapure water, in a confined volume (mixing region). During mixing, the hydrophobic QDs and copolymers precipitate out of the newly formed solution due to the presence of water which instantly imposes a supersaturation condition for the dissolved hydrophobic species. Simultaneously, the hydrophobic segments of the polymers immediately bind to isolated QDs and QD aggregates to form micelles where the hydrophilic blocks extend into the aqueous solution. The mixing time is required to be much shorter than the micellization time of block copolymer to realize flash nanoprecipitation. The design of the CIJM and MIVM units meet this requirement given adequate flow rates.26,28
Table 1 shows the effect of solvent-to-nonsolvent ratio and copolymer type on particle sizes. Decreasing solvent-to-nonsolvent ratio reduces the solubility of hydrophobic QDs. As a result, the supersaturation value is increased in the solvent mixture and therefore the particle size is decreased.39 The MIVM is used to generate the smallest particles due to the need for a high ratio of nonsolvent to solvent. This condition requires different flow rates of the inlet streams. The block copolymer composition and molecular weight can also affect the particle size owing to variable length of the hydrophilic and hydrophobic blocks. Pluronic F127 (structure is shown in Fig. 6 middle) is a difunctional block copolymer surfactant terminating in primary hydroxyl groups. Relatively nontoxic, it has recently been found to be temperature responsive and effective for drug and gene delivery.40 The pentablock copolymer (structure is shown in Fig. 6 top) used in this study is a unique functionalized derivative of Pluronic F127. It has triblock Pluronic F127 as its core where symmetric terminal blocks consist of poly(diethylaminoethyl methacrylate) (PDEAEM). The tertiary amine groups of PDEAEM are functional cationic segments that confer hydrophilicity at neutral pH and have been used previously in gene delivery experiments to electrostatically condense DNA and facilitate DNA release from endosomes post-uptake via the proton sponge effect.41 Poly(styrene-b-ethylene glycol) (PS-b-PEG, structure is shown in Fig. 6 bottom) is a simple diblock copolymer which has polystyrene as the hydrophobic block and polyethylene glycol as the hydrophilic block; likewise, this polymer also has low toxicity and is compatible with biological applications. These three varieties of block copolymers are not only capable of stabilizing hydrophobic QDs in water, but also potentially serve as carriers to deliver QDs into cells.
Table 1 Parametric effect on particle size
| |
PS(9.5k)-PEG(5k) (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1 NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
PS(9.5k)-PEG(5k) (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1 NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) 9) |
Pluronic F127 (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1 NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) 9) |
| Polydispersity |
0.245 |
0.024 |
0.086 |
| Z average size (nm) |
304 |
278.1 |
183.8 |
 |
| | Fig. 6 Chemical structure of pentablock copolymer (top); Pluronic F127 (middle); poly(styrene-b-ethylene glycol) (PS-PEG) (bottom). | |
3.2 Mixer effect on pentablock micelles
The pentablock copolymer used in this study forms micelles spontaneously in water at concentrations above the critical micelle concentration (CMC). Fig. 7 and Table 2 show that the size distribution of pentablock micelles (PBM) formed by the MIVM (Fig. 7) middle is much narrower than micelle size distributions formed without the mixer (Fig. 7) top, highlighting an advantage of using this system. However, after six hours the PDI of PBM increased substantially due to the reorganization of micelles as shown in Fig. 7 bottom. When mixed with hydrophobic QDs, the stability of QD-embedded PBM was greatly improved (Fig. 8). The hydrophobic QDs are thought to behave as stabilizing agents that maintain the structural integrity of the micelles.
 |
| | Fig. 7 Effect of MIVM on pentablock copolymer micelle size (Encapsulation Method I). Dissolved pentablock in water (no mixer, top). Forming pentablock micelles (PBM) using the mixer (middle). Pentablock micelles formed by the MIVM after 6 hours (bottom). | |
Table 2 Mixer effect on pentablock
| |
PBM (w/o mixer) |
PBM (w/mixer) |
PBM (w/mixer) 6 h |
| PDI |
1.00 |
0.304 |
0.511 |
| Z average size (nm) |
1887 |
251 |
227.6 |
 |
| | Fig. 8 QDs effect on pentablock micelle (Encapsulation Method I). Size distribution of QD-embedded PBM (PDI = 0.113) (top). Size distribution of QD-embedded PBM after one day (PDI = 0.078) (bottom). | |
3.3 Encapsulation Methods I–III
In order to obtain a wide range of nanoparticle sizes, we designed three different encapsulation methods. The first method uses the MIVM to obtain the smallest particles (diameters < 300 nm). Because it allows different momenta (i.e., flow rates) in each stream, a higher ratio of nonsolvent to solvent can be used. Although a lower ratio of nonsolvent to solvent leads to larger particles, there is an upper size limit around 200 nm. The small mixer volume and associated residence time of the MIVM significantly limits the size of QD aggregates and encapsulated particles. This limitation led to the second method which uses the CIJM and MIVM in series where a short tubing segment connects the two mixers. Equal volumes of THF (containing QDs and water), were pumped into the CIJM to form aggregates where they were subsequently sent to the MIVM for stabilization by block copolymer. The tubing connecting the two mixers was kept as short as possible to limit exposure to non-turbulent flow and suppress non-ideal aggregation conditions between mixer stages. This method provides QDs additional time to form aggregates, and consequently leads to larger particles following stabilization by polymers. As expected, increasing the tubing length between mixers increased the particle size as Table 3 shows, however this also increased the PDI. It therefore is not an ideal method for generating larger diameter particles (∼500 nm) unless the inevitably increased polydispersity is not a concern. Interestingly, it was discovered that the addition of hydrophobic homopolymer (polystyrene) into the nonsolvent stream can not only increase the mean particle size but also preserve narrow size distributions (Table 3). In general, longer hydrophobic chains resulted in larger particles.
Table 3 Size of QD-embedded nanoparticles using Encapsulation Method II. Each run in this table used PS (9.5k)-b-PEG (5k), solvent![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nonsolvent = 1
nonsolvent = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (CIJM)
1 (CIJM)
| |
PS-PEG/QD |
PS-PEG/QD (longer tubing connector) |
PS-PEG/QD/26k PS |
PS-PEG/QD/98k PS |
| PDI |
0.247 |
0.442 |
0.089 |
0.124 |
| Z average size (nm) |
322.2 |
448.9 |
314.1 |
533.9 |
The second encapsulation method was used to produce particle sizes up to 500 nm. To allow QDs an even longer aggregation time, a third method was used. Rather than using short tubing to connect the two mixers, the QD aggregates exiting the CIJM was first collected in a small vial and then pumped into the MIVM after a prescribed delay time (5–10 minutes). The precise time was determined beforehand by measuring the size of QD aggregates formed by the CIJM mixer at different time points. The variable delay time is ultimately limited by Ostwald ripening effects which serve to deteriorate the narrow size distribution. Critical parameters that led to larger particles sizes including equal amounts of solvent and nonsolvent and 98 kDa added polystyrene homopolymer were applied using this method. Under these optimized conditions, particles having mean diameters up to 800 nm were achievable using this approach (Fig. 9). Similar mixer effects were observed for all the polymers used in this work.
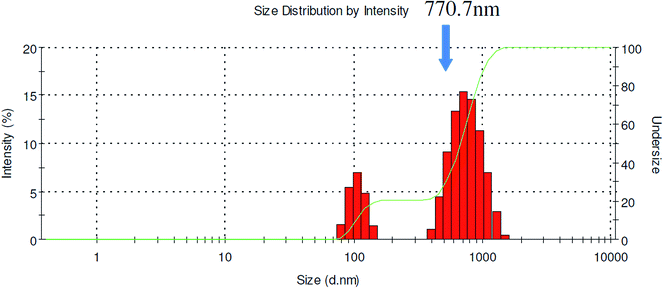 |
| | Fig. 9 Size distribution of QD-embedded nanoparticles using Encapsulation Method III (PDI = 0.445). | |
3.4 Characteristics of QD-embedded nanoparticles
One potential concern of this method is that QDs can aggregate inside micelles in such a way that would induce undesirable effects such as self-quenching, spectral broadening, and wavelength shifting.9 Fig. 10 shows the comparison of emission spectra of free QDs in THF and QD-embedded nanoparticles (Pluronic F127) in water. The photoluminescence (PL) of QDs decreased only slightly after being stabilized in Pluronic F127 copolymer, which indicates that individual QDs maintain sufficiently large interparticle distances due to the hydrophobic block of the copolymer. More surprisingly, the emission spectrum of QD-embedded nanoparticles was even narrower than that of freely dispersed QDs in solution. Another interesting finding is that the quenching effect can be greatly reduced by incorporating homopolymer into the polymer micelles with QDs; the molecular weight of the added homopolymer was inversely related to the observed quenching. As Fig. 10 shows, the PL intensity of QDs in the presence of 13 kDa polystyrene exhibited less quenching than particles having 5 kDa polystyrene. Regardless, both homopolymers were capable of reducing quenching because their association with QDs isolates them within the hydrophobic core of the micelle. This is demonstrated by size measurements as shown in Table 4. Incorporating QDs with homopolymer increases the size of the nanoparticles which is consistent with the hypothesis above where QDs have an increased average interparticle separation distance.
 |
| | Fig. 10 Comparison of emission spectra of free QDs in THF, QD-embedded nanoparticles using Pluronic F127 (QD-F127), and particles with homopolymer (5 kDa, 13 kDa polystyrene) in water. | |
Table 4 Size of QD-embedded nanoparticles using Encapsulation Method I. Each run in this table used Pluronic F127, solvent![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nonsolvent = 1
nonsolvent = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (MIVM)
9 (MIVM)
| |
F127/QD |
F127/QD/5k PS |
F127/QD/13k PS |
| PDI |
0.196 |
0.132 |
0.140 |
| Z average size (nm) |
138.0 |
170.5 |
175.2 |
Another common concern with water-soluble QDs is their pH stability, especially in acidic conditions. Most reported methods for generating water-soluble QDs do no produce nanoparticles that are stable in acidic solutions which limits their suitability for certain biological applications. In the case of our QD-embedded polymeric nanoparticles, the stabilizing block copolymer offers QDs improved solubility and pH stability (Fig. 11). These nanoparticles show excellent stability over a wide range of pH values. As Fig. 11 shows, even in a pH 3 buffer, QD luminescence is preserved. The superior pH stability should be attributed to Pluronic F127's nonionic feature.
 |
| | Fig. 11 Emission spectra of QD-embedded nanoparticles (Pluronic F127) in various pH buffer solutions. | |
One of the most appealing applications of QDs is as an energy donor for fluorescence resonance energy transfer (FRET) applications. QD can act as energy donor and once QD is close to its acceptor, its fluorescence will be quenched. Due to the high surface area-to-volume ratio of QDs, multiple biomolecules may be conjugated to a single QD which can greatly increase the overall FRET efficiency.42 Although hydrophobic QDs are well-protected by copolymer, they are still apparently accessible to biomolecules such as peptides dispersed in solution. Fig. 12 shows the composite emission spectra of QD-embedded nanoparticles (QD-F127) with an increasing number of Cy5 labeled peptide per QD. As shown, the decreased PL intensity of QDs and increasing Cy5 emission with an increasing number of peptides clearly demonstrates FRET between QDs and the Cy5-labeled peptide. The polymer offers the exceptional protect for QDs and preserve QDs FRET applications.
 |
| | Fig. 12 Composite emission spectra of QD-embedded nanoparticles (QD-F127) as a function of the number of Cy5-labeled peptides per QD (λex = 430 nm). | |
To evaluate the fluorescence intensity of a single particle, we examined samples using total internal reflection fluorescence microscopy (TIRFM) with a Nikon 60× TIRF objective (having a numerical aperture of 1.49). Excitation of the sample was achieved using a variable power diode laser system (Blue Sky Research, λex = 442 nm, 45 mW). The image was captured by a Photometrics CoolSNAP HQ2 CCD camera as shown in Fig. 13. The QD-encapsulated particles are monodisperse and extremely bright. No emission intermittency or “blinking” was observed in these samples indicating that most of the hydrophobic QDs (>99%) were successfully encapsulated into polymer micelles. A scanning electron microscopy (SEM) image shows the shape of these QD-embedded nanoparticles (Fig. 14), however some morphological changes from the native solution are expected due to the processing steps required for SEM. Transmission electron microscopy (TEM) could be used to image the embedded QDs due to their high contrast, however it is difficult to determine the actual number of QDs due to projection of the three-dimensional structure to a two-dimensional image.
 |
| | Fig. 13 False-color fluorescence image of QD-embedded polymeric nanoparticles (Pluronic F127 and 605 nm emitting CdSe–ZnS QDs) acquired using total internal reflection fluorescence microscopy (TIRFM) at 100× magnification. Scale bar = 1 μm. | |
 |
| | Fig. 14 SEM image of QD-embedded nanoparticles (poly(styrene-b-ethylene glycol)). | |
One of the most appealing applications for these QD-encapsulated nanoparticles is optical barcoding, which exploits the superior optical properties of QDs.9 As a proof of concept, we generated polymer particles via flash nanoprecipitation having roughly the same ratio of green-to-red QD peak signal intensity (as measured by a fluorimeter) adjusting for differences in extinction coefficient at the excitation wavelength (488 nm) and fluorescence quantum yield achieved through trial-and-error. These particles were then adsorbed from an aqueous solution onto a clean glass coverslip and imaged using TIRF microscopy as excited by the 488 nm line of an Ar-ion laser. Fig. 15 (top) reveals relatively similar spot intensities for individual particles as imaged using corresponding green and red emission filters. Small intensity differences were observed for co-localized spots in the green and red channels which were attributed differing optical filter bandwidths. In an ideal case, the filters would be optimized for the two signals and have equivalent intensities which are consistent with the ensemble measurements from the fluorimeter. A composite overlay of the green and red signals is shown in the Fig. 15 (bottom) where overlaid spots appear yellow indicating location registry between the channels. These images show the potential of this technique for generating a wide range of optically barcoded particles where the signals between particles within a particular population are consistent. However, a more thorough quantitative analysis is required to determine the variance in signal within the population. In our future work, we will use laser scanning confocal microscopy (LSCM) to generate a more rigorous quantitative analysis of the colloidal barcodes and determine the number of achievable orthogonal channels.
 |
| | Fig. 15 Images of polymer microparticles (∼250 nm mean diameter, 100× magnification) containing a mixture of green and red emitting TOP/TOPO-capped CdSe–ZnS QDs. Images were taken in TIRF mode using 488 nm excitation and relevant filter sets. The exposure time for each channel was identical at 300 ms. green channel (top), red channel (middle), (c) composite false color overlay of the green and red channel (co-localized spots appear in yellow) (bottom). | |
4 Conclusions
With the aid of a micromixer system, we have demonstrated a new and simple method that uses flash nanoprecipitation to generate QD-embedded nanoparticles having tunable mean size (∼90–800 nm) and narrow size distributions. We have examined numerous parameters including solvent-to-nonsolvent ratio, block copolymer composition and size, and homopolymer additives which collectively contribute to the final particle size distribution. We were able to preserve the initial bright photoluminescence of QDs following internalization through the addition of hydrophobic homopolymers. The nanoparticles produced by this micromixer-based technique are monodisperse, stable over a wide range of pH, and capable of FRET interactions upon exposure to dye-labeled molecules in solution. The block copolymers used to incorporate QDs are all nontoxic, biocompatible, and can have biofunctional surface functional groups which make this method appealing for biological applications.
To further extend the size range (mean diameters larger than 1 μm), we will likely have to slow the initial QD aggregation rate until they can be stabilized by copolymer condensation. Therefore, our future work will examine the QD surface ligand chemistry effect on particle size. The change of QD hydrophobicity will lead to variations in the supersaturation condition, which has a pronounced effect on the final size of QD-embedded nanoparticles.
Acknowledgements
We acknowledge financial support from startup funds at Iowa State University. Thanks to Dr Christopher Macosko (University of Minnesota, Dept. of Chemical Engineering & Material Science) and his group for kindly supplying the polystyrene homopolymers and offering helpful suggestions. We also thank the following colleagues at Iowa State University, Department of Chemical and Biological Engineering for their invaluable help with this project: Dr Rodney Fox for providing access to the micromixers, Dr Surya Mallapragada for providing Pluronic F127 and pentablock copolymers, Dr Eric Cochran for providing 98 kDa polystyrene homopolymers, and Dr Ian Schneider for providing imaging support.
References
- Z. Kaul, T. Yaguchi, S. C. Kaul, T. Hirano, R. Wadhwa and K. Taira, Cell Res., 2003, 13, 503–507 CrossRef PubMed
 .
. - D. Maysinger, M. Behrendt, M. Lalancette-Hbert and J. Kriz, Nano Lett., 2007, 7, 2513–2520 CrossRef CAS PubMed
 .
. - E. A. Jares-Erijman and T. M. Jovin, Nat. Biotechnol., 2003, 21, 1387–1395 CrossRef CAS PubMed
 .
. - X. Huang, L. Li, H. Qian, C. Dong and J. Ren, Angew. Chem., Int. Ed., 2006, 118, 5264–5267 CrossRef
 .
. - E. R. Goldman, A. R. Clapp, G. P. Anderson, H. T. Uyeda, J. M. Mauro, I. L. Medintz and H. Mattoussi, Anal. Chem., 2004, 76, 684688 CrossRef PubMed
 .
. - V. V. Klimov, M. Ducloy and V. S. Letokhov, Phys. Rev. A, 1999, 59, 2996 CrossRef CAS
 .
. - J. N. Cha, M. H. Bartl, M. S. Wong, A. Popitsch, T. J. Deming and G. D. Stucky, Nano Lett., 2003, 3, 907–911 CrossRef CAS
 .
. - M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS
 .
. - M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631635 Search PubMed
 .
. - A. C. C. Esteves, A. Barros-Timmons, T. Monteiro and T. Trindade, J. Nanosci. Nanotechnol., 2005, 5, 766–771 CrossRef CAS PubMed
 .
. - M. Peres, L. C. Costa, A. Neves, M. J. Soares, T. Monteiro, A. C. Esteves, A. Barros-Timmons, T. Trindade, A. Kholkin and E. Alves, Nanotechnology, 2005, 16, 1969–1973 CrossRef CAS
 .
. - W. Sheng, S. Kim, J. Lee, S. Kim, K. Jensen and M. G. Bawendi, Langmuir, 2006, 22, 3782–3790 CrossRef CAS PubMed
 .
. - P. O'Brien, S. S. Cummins, D. Darcy, A. Dearden, O. Masala, N. L. Pickett, S. Ryley and A. J. Sutherland, Chem. Commun., 2003, 2532 RSC
 .
. - M. Kuang, D. Wang, H. Bao, M. Gao, H. Mhwald and M. Jiang, Adv. Mater., 2005, 17, 267–270 CrossRef CAS
 .
. - J. Li, B. Liu and J. Li, Langmuir, 2006, 22, 528–531 CrossRef CAS PubMed
 .
. - U. Hasegawa, S. M. Nomura, S. C. Kaul, T. Hirano and K. Akiyoshi, Biochem. Biophys. Res. Commun., 2005, 331, 917–921 CrossRef CAS PubMed
 .
. - D. Wang, A. L. Rogach and F. Caruso, Nano Lett., 2002, 2, 857–861 CrossRef CAS
 .
. - M. Moffitt, H. Vali and A. Eisenberg, Chem. Mater., 1998, 10, 1021–1028 CrossRef CAS
 .
. - H. Yusuf, W. Kim, D. H. Lee, Y. Guo and M. G. Moffitt, Langmuir, 2007, 23, 868–878 CrossRef CAS PubMed
 .
. - Y. Chen and Z. Rosenzweig, Nano Lett., 2002, 2, 1299–1302 CrossRef CAS
 .
. - A. C. Kamps, B. L. Sanchez-Gaytan, R. J. Hickey, N. Clarke, M. Fryd and S. Park, Langmuir, 2010, 26, 14345–14350 CrossRef CAS PubMed
 .
. - Y. Yang and M. Y. Gao, Adv. Mater., 2005, 17, 2354–2357 CrossRef CAS
 .
. - D. K. Yi, S. T. Selvan, S. S. Lee, G. C. Papaefthymiou, D. Kundaliya and J. Y. Ying, J. Am. Chem. Soc., 2005, 127, 4990–4991 CrossRef CAS PubMed
 .
. - S. T. Selvan, T. T. Tan and J. Y. Ying, Adv. Mater., 2005, 17, 1620–1625 CrossRef CAS
 .
. - J. Yang, S. R. Dave and X. Gao, J. Am. Chem. Soc., 2008, 130, 5286–5292 CrossRef CAS PubMed
 .
. - B. K. Johnson and R. K. Prud'homme, Aust. J. Chem., 2003, 56, 1021–1024 CrossRef CAS
 .
. - B. K. Johnson and R. K. Prud'homme, AIChE J., 2003, 49, 2264–2282 CrossRef CAS
 .
. - Y. Liu, C. Cheng, Y. Liu, R. K. Prud'homme and R. O. Fox, Chem. Eng. Sci., 2008, 63, 2829–2842 CrossRef CAS PubMed
 .
. - A. R. Clapp, E. R. Goldman and H. Mattoussi, Nat. Protoc., 2006, 1, 12581266 CrossRef PubMed
 .
. - Y. Zhang, A. M. Schnoes and A. R. Clapp, ACS Appl. Mater. Interfaces, 2010, 2, 3384–3395 CAS
 .
. - B. J. Berne and R. Pecora, Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics, Dover Publications, Unabridged edn, 2000 Search PubMed
 .
. - C. F. Bohren and D. R. Huffman, Absorption and Scattering of Light by Small Particles, Wiley-VCH, 1998 Search PubMed
 .
. - B. J. Frisken, Appl. Opt., 2001, 40, 4087–4091 CrossRef CAS
 .
. - D. E. Koppel, J. Chem. Phys., 1972, 57, 4814 CrossRef CAS PubMed
 .
. - W. B. Russel, D. A. Saville and W. R. Schowalter, Colloidal Dispersions, Cambridge University Press, 1992 Search PubMed
 .
. - Y. Liu, PhD thesis, Princeton University, 2007
 .
. - Y. Liu and R. O. Fox, AIChE J., 2006, 52, 731–744 CrossRef CAS
 .
. - Z. Zhu, J. L. Anacker, S. Ji, T. R. Hoye, C. W. Macosko and R. K. Prud'homme, Langmuir, 2007, 23, 10499–10504 CrossRef CAS PubMed
 .
. - W. Liu, H. S. Choi, J. P. Zimmer, E. Tanaka, J. V. Frangioni and M. Bawendi, J. Am. Chem. Soc., 2007, 129, 14530–14531 CrossRef CAS PubMed
 .
. - A. V. Kabanov, E. V. Batrakova and V. Y. Alakhov, J. Controlled Release, 2002, 82, 189–212 CrossRef CAS
 .
. - B. Zhang, M. Kanapathipillai, P. Bisso and S. Mallapragada, Pharm. Res., 2009, 26, 700–713 CrossRef CAS PubMed
 .
. - A. R. Clapp, I. L. Medintz, J. M. Mauro, B. R. Fisher, M. G. Bawendi and H. Mattoussi, J. Am. Chem. Soc., 2004, 126, 301–310 CrossRef CAS PubMed
 .
.
|
| This journal is © The Royal Society of Chemistry 2014 |
Click here to see how this site uses Cookies. View our privacy policy here. 

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 solvent-to-nonsolvent ratio (S
9 solvent-to-nonsolvent ratio (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS) case, two streams (streams 1 and 3) delivered a total of 45 mL of water (nonsolvent, NS), stream 2 flowed 5 mL of THF (solvent, S) with 20 mg of dissolved QDs, and stream 4 flowed 5 mL of THF with 30 mg of dissolved block copolymer. The ratio of solvent to nonsolvent was 1
NS) case, two streams (streams 1 and 3) delivered a total of 45 mL of water (nonsolvent, NS), stream 2 flowed 5 mL of THF (solvent, S) with 20 mg of dissolved QDs, and stream 4 flowed 5 mL of THF with 30 mg of dissolved block copolymer. The ratio of solvent to nonsolvent was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (S
9 (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1
NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) to ensure supersaturation conditions and rapid nanoprecipitation. The flow rates of streams 1 and 3 were 70 mL min−1, while that of streams 2 and 4 were 7.8 mL min−1. In the 1
9) to ensure supersaturation conditions and rapid nanoprecipitation. The flow rates of streams 1 and 3 were 70 mL min−1, while that of streams 2 and 4 were 7.8 mL min−1. In the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 S
1 S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS case, the delivered volumes were matched at 5 mL in all four streams. The flow rate of each stream was controlled by a syringe pump. All four streams were set to start and stop simultaneously. The product stream leaving the MIVM was collected in a vial where samples were later characterized by a Malvern Zetasizer Nano-ZS90.
NS case, the delivered volumes were matched at 5 mL in all four streams. The flow rate of each stream was controlled by a syringe pump. All four streams were set to start and stop simultaneously. The product stream leaving the MIVM was collected in a vial where samples were later characterized by a Malvern Zetasizer Nano-ZS90.






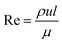
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1
NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)
1)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1
NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9)
9)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NS = 1
NS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9)
9)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nonsolvent = 1
nonsolvent = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (CIJM)
1 (CIJM)

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nonsolvent = 1
nonsolvent = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (MIVM)
9 (MIVM)


.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.






