Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanopatterns of polymer brushes for understanding protein adsorption on the nanoscale†
Xiaowen
Wang
ab,
Rüdiger
Berger
a,
Jagoba Iturri
Ramos
a,
Tao
Wang
b,
Kaloian
Koynov
a,
Guangming
Liu
*b,
Hans-Jürgen
Butt
a and
Si
Wu
*a
aMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany. E-mail: wusi@mpip-mainz.mpg.de
bHefei National Laboratory for Physical Sciences at the Microscale, Department of Chemical Physics, University of Science and Technology of China, Hefei 230026, P. R. China. E-mail: gml@ustc.edu.cn
First published on 5th September 2014
Abstract
We present fabrication of patterned poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) brushes with sub-100 nm features over large areas. The patterned polymer brushes are fabricated by a combination of block copolymer micelle lithography and surface-initiated atom transfer radical polymerization. The PDMAEMA brushes are neutralized and collapsed at pH 9, and positively charged and swollen at pH 4. The protein adsorption and desorption on the patterned PDMAEMA brushes are studied by laser scanning confocal microscopy, atomic force microscopy (AFM), and quartz crystal microbalance with dissipation (QCM-D). In 1 mM NaCl solution at pH 5.8, the patterned brushes take up bovine serum albumin (BSA, isoelectric point ∼4.8) via electrostatic interactions. BSA adsorbs both inside the brushes and at the outer edge of the brushes. BSA at the outer edge of the brushes is released by rinsing the brushes with 1 M NaCl solutions at pH 4 and 9. Part of the absorbed BSA remains trapped inside the brushes, resulting in an increase of their volume. The regular sub-100 nm features of the patterned PDMAEMA brushes allowed us to directly visualize protein adsorption/desorption by AFM on a nanoscale. The large area of the patterned brushes allowed us to collect statistical results of the nanostructures by QCM-D.
Introduction
Polymer brushes are layers of polymer chains that are tethered to a surface with one chain ending.1 Polymer brushes can selectively bind proteins, which may open applications in protein immobilization, purification, and analysis.2 Understanding protein adsorption/desorption on polymer brushes will help to delineate their performance for such applications. It was demonstrated that controlling the environmental pH is a practical way to manipulate protein adsorption/desorption on pH-responsive polymer brushes.3–8 Protein adsorption/desorption on pH-responsive polymer brushes has been studied using quartz crystal microbalance with dissipation (QCM-D),8 surface plasmon resonance (SPR) spectroscopy,4 and optical reflectometry.6,7 However, these methods lack spatial resolution, which only provides limited information for protein adsorption/desorption on polymer brushes. Direct imaging protein adsorption/desorption on polymer brushes at the nano scale can give us more insights into the performance of polymer brushes as protein binders.Patterned polymer brushes were used as model systems to understand wettability, adhesion, and adsorption on surfaces.9–11 Patterned polymer brushes with small feature sizes may enable us to directly image protein adsorption/desorption on polymer brushes at the nano scale. In a recent review, Chen et al. summarized that patterned polymer brushes are fabricated by combination of various lithography methods and surface-initiated polymerizations.9 The smallest feature size of patterned polymer brushes fabricated by conventional photolithography is in the order of few hundred nanometers caused by the diffraction limit of light.9 The feature size of patterned polymer brushes fabricated by micro contact printing is in the order of microns.9 Electron-beam lithography and scanning probe lithography can produce patterned polymer brushes with sub-100 nm features.9 Due to the low throughput of electron-beam lithography and scanning probe lithography, the areas of patterned polymer brushes are usually only up to ∼100 μm2. This area is too small for some standard characterization techniques such as gravimetry by quartz crystal microbalance. Cell adhesion study is also not applicable on patterns with small areas because T cells require a certain density of continuously nanopatterns for spreading to occur and a large-size extracellular matrix is needed to collect statistical results.12,13 Scanning probe lithography using tip arrays can increase the throughput of patterned polymer brushes.14 The problem of scanning probe lithography using tip arrays is that patterning will fail if substrates are uneven or tips in tip arrays are not perfectly aligned.14 Recently, block copolymer lithography based on phase separation of block copolymers was combined with surface-initiated polymerizations to fabricate patterned polymer brushes.15,16 The fabricated patterned polymer brushes may not fully replicate ordered structures of block copolymer templates.15 This may occur because the pattern transfer processes for patterns with sub-100 nm features are difficult to achieve. Up until now, it is still a challenge to fabricate patterned polymer brushes with sub-100 nm features over large areas.
Our goals are (i) the demonstration of a new method for fabricating patterned polymer brushes with sub-100 nm features over large areas and (ii) the use of these novel fabricated patterned polymer brushes to study protein adsorption. The pH-responsive patterned poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) brushes were fabricated by combination of block copolymer micelle lithography and surface-initiated atom transfer radical polymerization (SI-ATRP). Our polymer brushes are grafted onto patterned Au nanoparticles via the “grafting from” method. In contrast to the “grafting to” method,12,13,17,18 polymer brushes with a high grafting density can be synthesized using the “grafting from” method.1,19–23 To the best of our knowledge, fabrication of patterned polymer brushes by SI-ATRP from patterned Au nanoparticles prepared by block copolymer micelle lithography has not been reported yet. The regular sub-100 nm features of the patterned PDMAEMA brushes allowed us to directly visualize protein adsorption/desorption on the patterned brushes by atomic force microscopy (AFM) on a nano scale. The large area of the patterned PDMAEMA brushes allowed us to use QCM-D to collect statistical results of the nanostructures.
Experimental section
Materials
Polystyrene-block-poly(2-vinylpyridine) (PS-b-P2VP, Mn = 55-b-50 kg mol−1 and PDI = 1.05) and ω-trimethoxysilane terminated PEG methyl ether (Mn = 0.35 kg mol−1 and PDI = 1.10) were purchased from Polymer Source. Bovine serum albumin (BSA) labeled with fluorescent Texas Red was purchased from Life Technologies and used without further purification. The ATRP initiator ω-mercaptoundecyl bromoisobutyrate was synthesized according to the literature.24 All other chemicals were purchased from Sigma-Aldrich.Sample preparation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The micelle solution was stirred for 1 day to allow it to reach equilibrium. Dip-coating of clean quartz slides or quartz crystals into the micelle solution with a velocity of 12 mm min−1 produced a monolayer of micelles. Patterned Au nanoparticles were obtained by treating the monolayer with oxygen plasma (5 sccm, 15 W, 15 min) and annealing in H2 (200 °C, 1.5 h).
2. The micelle solution was stirred for 1 day to allow it to reach equilibrium. Dip-coating of clean quartz slides or quartz crystals into the micelle solution with a velocity of 12 mm min−1 produced a monolayer of micelles. Patterned Au nanoparticles were obtained by treating the monolayer with oxygen plasma (5 sccm, 15 W, 15 min) and annealing in H2 (200 °C, 1.5 h).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). After stirring and bubbling with Ar for 30 min, CuBr (0.2 mmol, 0.0285 g) was added under Ar. The mixture was stirred for another 30 min. Then, the initiator-modified substrates were placed inside the flask under Ar. The polymerization reaction was at 25 °C for 24 h. After polymerization, the substrates were washed with water and methanol. The space between nanoparticles was passivated by PEG using a literature method with minor modifications.12,13 In brief, PEG passivation is conducted by immersing substrates in an anhydrous toluene solution (30 mL) of ω-trimethoxysilane terminated PEG methyl ether (0.07 g) and triethylamine as the catalyst. The substrate was placed in the above solution under Ar at 40 °C for 4 h. Then, the substrate was washed with toluene and ethanol.
1 v/v). After stirring and bubbling with Ar for 30 min, CuBr (0.2 mmol, 0.0285 g) was added under Ar. The mixture was stirred for another 30 min. Then, the initiator-modified substrates were placed inside the flask under Ar. The polymerization reaction was at 25 °C for 24 h. After polymerization, the substrates were washed with water and methanol. The space between nanoparticles was passivated by PEG using a literature method with minor modifications.12,13 In brief, PEG passivation is conducted by immersing substrates in an anhydrous toluene solution (30 mL) of ω-trimethoxysilane terminated PEG methyl ether (0.07 g) and triethylamine as the catalyst. The substrate was placed in the above solution under Ar at 40 °C for 4 h. Then, the substrate was washed with toluene and ethanol.
Characterization
X-ray photoelectron spectroscopy (XPS) measurements were conducted on an ESCALAB-250 spectrometer with a monochromatic Al Kα X-ray source (hν = 1486.6 eV). Atomic force microscopy (AFM) images were obtained on a Dimension 3100 system using tapping mode. A silicon cantilever (OMCL AC 160 TN-W2, spring constant ∼42 N m−1, resonance frequency ∼300 kHz) was used. Scanning electron microscopy (SEM) images were obtained on a LEO Gemini 1530 system. Quartz crystal microbalance with dissipation (QCM-D) experiments were conducted on a Q-sense E1 system. Quartz crystals with patterned PDMAEMA brushes were used as QCM sensors. Laser scanning confocal microscopy experiments were performed on a commercial setup (Carl Zeiss, Jena, Germany) consisting of the module LSM 510 and an inverted microscope model Axiovert 200 using a C-Apochromat 40× water immersion objective (Carl Zeiss, Jena, Germany) with numerical aperture (NA) of 1.2. The excitation was done with the 543 nm line of a 1 mW HeNe laser fiber coupled to the LSM510 module. The fluorescent images were recorded using a LP 560 long-pass emission filter.Results and discussion
Patterned polymer brushes
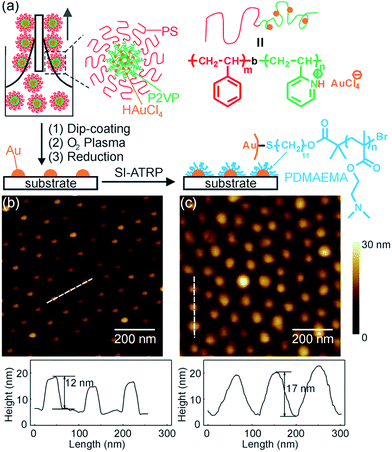
To fabricate patterned PDMAEMA brushes (Fig. 1a), first, a micelle solution of PS-b-P2VP in toluene was loaded with HAuCl4. HAuCl4 selectively bound to the P2VP block via protonation. Then, a close-packed monolayer of micelles on a quartz substrate was fabricated by dip-coating. PS-b-P2VP was removed, and patterned Au nanoparticles were formed when the monolayer was exposed to oxygen plasma and annealed in a hydrogen atmosphere.18,25,26 This method can generate patterned Au nanoparticles on a wafer scale.18,25,26 The Au nanoparticles which were prepared using the optimized plasma and annealing conditions are stable and difficult to remove from the quartz substrate even by sonication.18 The stability, large area and small feature size of the patterned Au nanoparticles make them interesting templates for fabricating large-area and high-resolution patterns of polymer brushes. However, fabrication of patterned polymer brushes by SI-ATRP from patterned Au nanoparticles prepared by block copolymer micelle lithography has not been reported. To fabricate such patterned polymer brushes, an ATRP initiator ω-mercaptoundecyl bromoisobutyrate was selectively anchored to the patterned Au nanoparticles. Finally, patterned PDMAEMA brushes were grafted from the patterned Au nanoparticles by SI-ATRP (Fig. 1a).Block copolymer micelle lithography generated a quasi-hexagonal pattern of Au nanoparticles on the quartz substrate (Fig. 1b). The average distance of neighboring Au nanoparticles was 91 nm (Fig. S1†). The average height of Au nanoparticles measured by AFM in tapping mode in air was 10.3 ± 2.4 nm (Fig. 1b and S2†). After grafting PDMAEMA brushes, the average height of the nanoparticles increased to 18.4 ± 4.5 nm (Fig. 1c and S2†), indicating that Au nanoparticles were coated with an approximately 8 nm polymer layer. We stochastically analyzed the morphology of patterned PDMAEMA brushes at different positions on the same substrate (Fig. S3†). The similar morphology of patterned PDMAEMA brushes at randomly different positions indicates that PDMAEMA brushes were patterned over a large area (Fig. S3†).
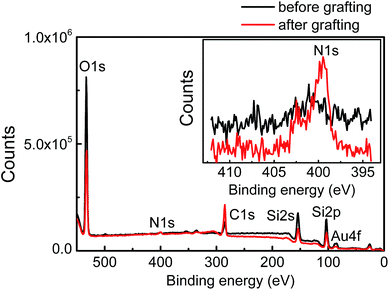
Surface compositions of patterned Au nanoparticles before and after grafting PDMAEMA were studied by X-ray photoelectron spectroscopy (XPS, Fig. 2). After PDMAEMA was grafted from the Au nanoparticles, the N 1s signal at ∼399 eV, which corresponds to the dimethylamino group, appeared. The intensity of Au 4f signals at ∼84.5 and ∼88.5 eV decreased. This measurement confirmed the presence of PDMAEMA on the surface.
pH responsiveness of the patterned polymer brushes
PDMAEMA is pH-responsive resulting from the protonation/deprotonation equilibrium of the dimethylamino groups (Fig. 3a).4,27 We studied pH response of patterned PDMAEMA brushes using QCM-D (Fig. 3b). To avoid the disturbance of acid–base reactions, we always used a pH 7 buffer solution to rinse the QCM cell when switching pH between 4 and 9. Frequency shift (Δf) and dissipation shift.(ΔD) increased when the pH was switched from 7 to 4. As pH decreased from 7 to 4, the extent of hydration of PDMAEMA increased due to the protonation of the dimethylamino groups. This hydration effect, which was observed on PDMAEMA brushes on a flat surface,28 will decrease Δf. However, for surfaces with nanostructures, Δf is influenced not only by the hydration/dehydration of polymer chains but also by the water trapped in surface nanostructures.29,30 The swelling of PDMEMA on the patterned surface with decreasing pH from 7 to 4 will lessen the surface roughness, which reduces the amount of water trapped by the surface nanostructures. Therefore, the effect of the nanostructures will increase Δf. The combined effects by hydration and surface nanostructures resulted in an increase of Δf with decreasing pH from 7 to 4 (Fig. 3b). This result indicates that the mass change induced by the reduction of trapped water in surface nanostructures dominates over that induced by the hydration of PDMAEMA. ΔD increased with decreasing pH from 7 to 4, indicating that more energy was dissipated in the swollen PDMAEMA layer via internal friction.
When pH changed from 7 to 9, Δf increased and ΔD decreased (Fig. 3b). These results imply that the deprotonation of dimethylamino groups caused dehydration of PDMAEMA and the weakening of the electrostatic repulsions between PDMAEMA chains cause a collapse of PDMAEMA chains. The collapse of PDMAEMA might generate a rougher surface, but the collapsed PDMAEMA chains are too hydrophobic to effectively trap water.31,32 Thus, the mass change of the patterned surface was dominated by the dehydration of PDMAEMA with increasing pH from 7 to 9. The cyclic measurements by QCM-D show that the pH response of the patterned brushes was reversible. The control experiment using patterned Au nanoparticles without PDMAEMA showed that both Δf and ΔD remained almost constant with varying pH (Fig. S4†), confirming that the pH response was induced by the variation of PDMAEMA brushes at different pH.
Protein adsorption
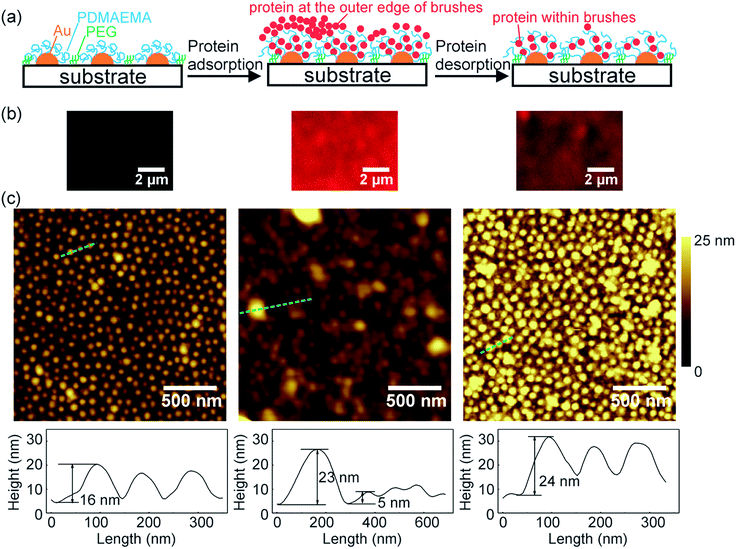
We studied protein adsorption and desorption on patterned PDMAEMA brushes (Fig. 4). To avoid protein adsorption on the substrate between the polymer pattern, the surface was modified with a protein resistant poly(ethylene glycol) (PEG) layer. Laser scanning confocal microscopy was used to study adsorption and desorption of fluorescence-labeled bovine serum albumin (BSA, isoelectric point ∼4.8) (Fig. 4b). The patterned brushes were non-fluorescent (Fig. 4b left). Fluorescence appeared only after immersion in BSA solution (0.1 mg mL−1, pH 5.8) for 1 hour and subsequent washing with a NaCl solution (1 mM, pH 5.8). This fluorescence indicates that BSA was adsorbed on the surface. The fluorescence intensity decreased to ∼47% of the original intensity after the sample was successively rinsed in NaCl solution (1 M, pH 4) and NaCl solution (1 M, pH 9) (Fig. 4b right and S5†), which indicates that ∼53% of the adsorbed proteins were released from the surface. In a control experiment, substrates without PDMAEMA showed nearly no protein adsorption (Fig. S6†), demonstrating that proteins were taken up by PDMAEMA brushes.Proteins may penetrate into polymer brushes or adsorb at the outer edge of brushes (Fig. 4a).3 We used AFM to study the locations of proteins adsorbed on the patterned PDMAEMA brushes (Fig. 4c). The average height of the nanostructures in the initial patterned brushes was 15.4 ± 3.5 nm (Fig. 4c left and S7†). After the brushes were exposed to BSA, the pattern became blurry (Fig. 4c middle). The average height of the nanostructures decreased to 5.5 ± 1.7 nm and some big aggregates appeared on the surface. These results indicate that some proteins on the outer edge of the brushes formed aggregates and merged initially separated nanostructures (model in Fig. 4a middle). After part of the adsorbed BSA was released from the surface again, an ordered pattern appeared (Fig. 4c right). However, the average height of the nanostructures increased to 19.5 ± 5.1 nm that is even higher than that of the nanostructures before protein adsorption (Fig. S7†). According to the laser scanning confocal microscopy images (Fig. 4b), ∼47% of the proteins were still on the surface after the desorption process. These results show that the residual proteins were located inside the brushes. Accumulation of proteins inside the brushes caused the volume expansion of the nanostructures.
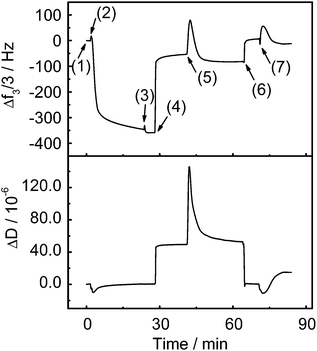
The kinetics of protein adsorption/desorption on the patterned PDMAEMA brushes were studied by QCM-D (Fig. 5). To obtain the baseline, the patterned brushes were exposed to NaCl solution (1 mM, pH 5.8, step (1)). When the patterned brush surfaces were exposed to BSA solution (0.1 mg mL−1, pH 5.8, step (2)), Δf decreased drastically within 3 min. It continued to decrease slowly in the following 20 min until saturation. This Δf decrease indicates a mass increase due to adsorbed proteins. There was only a slight change in ΔD during protein adsorption. The adsorption of BSA on the patterned brushes should lead to an increase of ΔD because the adsorbed BSA will cause more energy to be dissipated during the oscillation of the resonator. Nonetheless, the electrostatic complexation between the adsorbed BSA and the brushes would make the polymer layer more rigid as BSA and PDMAEMA are oppositely charged at pH 5.8. This effect will decrease ΔD. As a result, the combined effects may only lead to a slight change in ΔD during the protein adsorption process (Fig. 5). Additionally, the adsorbed proteins are stable upon rinsing with NaCl solution (1 mM, pH 5.8, step (3)).
To release protein, the patterned brushes were rinsed with NaCl solution (1 M, pH 4, setp (4)). As a consequence, both Δf and ΔD rapidly increased (Fig. 5). The increase in Δf indicates the desorption of some proteins. At pH 4, both BSA and PDMAEMA are positively charged. The high ionic strength in 1 M NaCl solution can screen localized electrostatic attractions and provide high-concentration Cl− as counter ions to exchange BSA from the brushes. Both pH and ionic strength should contribute to protein desorption.4,27 However, some strongly adsorbed proteins still stick to the brushes. This should be due to the electrostatic attractions between the positively charged brushes and the negatively charged patches on BSA because pH 4 is close to the isoelectric point of BSA (pH 4.8). These adsorbed proteins will form a swollen layer on the surface of the brushes as they are repelled by the brushes, which would increase ΔD. Then, the patterned brushes were rinsed with NaCl solution again (1 mM, pH 5.8, step (5)). To further release the adsorbed proteins, the patterned brushes were successively rinsed with NaCl solution (1 M, pH 9, step (6)) and NaCl solution (1 mM, pH 5.8, step (7)). PDMAEMA is neutral at pH 9 and no electrostatic attractions exist between the brushes and BSA at this pH. Consequently, the residual BSA molecules at the outer edge of the brushes were removed by rinsing with the NaCl solutions, as indicated by the further increase in Δf and decrease in ΔD. In a control experiment, patterned Au nanoparticles without PDMAEMA adsorbed hardly any protein (Fig. S8†).
Conclusions
By combination of block copolymer micelle lithography and SI-ATRP, we developed an efficient method to fabricate pH-responsive patterned polymer brushes with sub-100 nm features over large areas. The small feature size of the patterned brushes allowed us to visualize protein adsorption/desorption by AFM on a nano scale. The large area of the patterned brushes allowed us to collect statistical results of the nanostructures by QCM-D. Thus, the patterned polymer brushes could be used as a prototype system to understand fundamental questions regarding protein adsorption/desorption on a nano scale. We expect that large-area nanopatterns of polymer brushes with various stimuli-responsive properties and functionalities could be synthesized by our method using other monomers instead of DMAEMA.Acknowledgements
X.W. was supported by the joint program of MPG and CAS. We thank G. Glasser for SEM measurements, K. Bley and C. K. Weiss for oxygen plasma treatment, and R. Li and X. Feng for annealing samples in hydrogen atmosphere. This research was partly supported by National Natural Science Foundation of China (21374110 and 91127042).Notes and references
- R. Barbey, L. Lavanant, D. Paripovic, N. Schüwer, C. Sugnaux, S. Tugulu and H. A. Klok, Chem. Rev., 2009, 109, 5437–5527 CrossRef CAS PubMed.
- P. Jain, G. L. Baker and M. L. Bruening, Annu. Rev. Anal. Chem., 2009, 2, 387–408 CrossRef CAS PubMed.
- H. Kuroki, I. Tokarev and S. Minko, Annu. Rev. Mater. Res., 2012, 42, 343–372 CrossRef CAS.
- A. Kusumo, L. Bombalski, Q. Lin, K. Matyjaszewski, J. W. Schneider and R. D. Tilton, Langmuir, 2007, 23, 4448–4454 CrossRef CAS PubMed.
- Q. A. Yu, H. Chen, Y. X. Zhang, L. Yuan, T. L. Zhao, X. Li and H. W. Wang, Langmuir, 2010, 26, 17812–17815 CrossRef CAS PubMed.
- R. Zimmermann, T. Osaki, T. Kratzmuller, G. Gauglitz, S. S. Dukhin and C. Werner, Anal. Chem., 2006, 78, 5851–5857 CrossRef CAS PubMed.
- W. M. de Vos, P. M. Biesheuvel, A. de Keizer, J. M. Kleijn and M. A. C. Stuart, Langmuir, 2008, 24, 6575–6584 CrossRef CAS PubMed.
- E. Bittrich, K. B. Rodenhausen, K. J. Eichhorn, T. Hofmann, M. Schubert, M. Stamm and P. Uhlmann, Biointerphases, 2010, 5, 159–167 CrossRef PubMed.
- T. Chen, I. Amin and R. Jordan, Chem. Soc. Rev., 2012, 41, 3280–3296 RSC.
- M. A. C. Stuart, W. T. S. Huck, J. Genzer, M. Müller, C. Ober, M. Stamm, G. B. Sukhorukov, I. Szleifer, V. V. Tsukruk, M. Urban, F. Winnik, S. Zauscher, I. Luzinov and S. Minko, Nat. Mater., 2010, 9, 101–113 CrossRef PubMed.
- C. Schuh, S. Santer, O. Prucker and J. Rühe, Adv. Mater., 2009, 21, 4706–4710 CAS.
- J. Deeg, M. Axmann, J. Matic, A. Liapis, D. Depoil, J. Afrose, S. Curado, M. L. Dustin and J. P. Spatz, Nano Lett., 2013, 13, 5619–5626 CrossRef CAS PubMed.
- J. Matic, J. Deeg, A. Scheffold, I. Goldstein and J. P. Spatz, Nano Lett., 2013, 13, 5090–5097 CrossRef CAS PubMed.
- X. C. Zhou, Z. L. Liu, Z. Xie, X. Q. Liu and Z. J. Zheng, Small, 2012, 8, 3568–3572 CrossRef CAS PubMed.
- Y. Liu, H. Y. Hu, W. C. Ye, F. Zhou and J. C. Hao, J. Mater. Chem. C, 2013, 1, 902–907 RSC.
- D. P. Sweat, M. Kim, X. Yu, S. K. Schmitt, E. Han, J. W. Choi and P. Gopalan, Langmuir, 2013, 29, 12858–12865 CrossRef CAS PubMed.
- T. Lohmüller, S. Triffo, G. P. O'Donoghue, Q. Xu, M. P. Coyle and J. T. Groves, Nano Lett., 2011, 11, 4912–4918 CrossRef PubMed.
- A. C. Pearson, E. Pound, A. T. Woolley, M. R. Linford, J. N. Harb and R. C. Davis, Nano Lett., 2011, 11, 1981–1987 CrossRef CAS PubMed.
- G. N. Ju, M. J. Cheng, M. Xiao, J. M. Xu, K. Pan, X. Wang, Y. J. Zhang and F. Shi, Adv. Mater., 2013, 25, 2915–2919 CrossRef CAS PubMed.
- F. Krohm, H. Didzoleit, M. Schulze, C. Dietz, R. W. Stark, C. Hess, B. Stühn and A. Brunsen, Langmuir, 2014, 30, 369–379 CrossRef CAS PubMed.
- J. Elbert, F. Krohm, C. Rüttiger, S. Kienle, H. Didzoleit, B. N. Balzer, T. Hugel, B. Stühn, M. Gallei and A. Brunsen, Adv. Funct. Mater., 2014, 24, 1591–1601 CrossRef CAS.
- A. Farrukh, A. Akram, A. Ghaffar, S. Hanif, A. Hamid, H. Duran and B. Yameen, ACS Appl. Mater. Interfaces, 2013, 5, 3784–3793 CAS.
- B. Yameen and A. Farrukh, Chem.–Asian J., 2013, 8, 1736–1753 CrossRef CAS PubMed.
- H. W. Ma, M. Wells, T. P. Beebe and A. Chilkoti, Adv. Funct. Mater., 2006, 16, 640–648 CrossRef CAS.
- R. Glass, M. Möller and J. P. Spatz, Nanotechnology, 2003, 14, 1153–1160 CrossRef CAS.
- G. Kästle, H. G. Boyen, F. Weigl, G. Lengl, T. Herzog, P. Ziemann, S. Riethmüller, O. Mayer, C. Hartmann, J. P. Spatz, M. Möller, M. Ozawa, F. Banhart, M. G. Garnier and P. Oelhafen, Adv. Funct. Mater., 2003, 13, 853–861 CrossRef.
- S. Dai, P. Ravi and K. C. Tam, Soft Matter, 2008, 4, 435–449 RSC.
- X. W. Wang, G. M. Liu and G. Z. Zhang, Langmuir, 2011, 27, 9895–9901 CrossRef CAS PubMed.
- S. Wehner, K. Wondraczek, D. Johannsmann and A. Bund, Langmuir, 2004, 20, 2356–2360 CrossRef CAS.
- S. J. Martin, G. C. Frye, A. J. Ricco and S. D. Senturia, Anal. Chem., 1993, 65, 2910–2922 CrossRef CAS.
- K. A. Marx, Biomacromolecules, 2003, 4, 1099–1120 CrossRef CAS PubMed.
- B. Y. Du, E. Goubaidoulline and D. Johannsmann, Langmuir, 2004, 20, 10617–10624 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Statistical analyses, SEM images, laser scanning confocal microscopy images and additional QCM-D data. See DOI: 10.1039/c4ra07623k |
| This journal is © The Royal Society of Chemistry 2014 |