Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structural characterization of nanofiber silk produced by embiopterans (webspinners)†
J. Bennett
Addison
a,
Thomas M.
Osborn Popp
a,
Warner S.
Weber
a,
Janice S.
Edgerly
b,
Gregory P.
Holland
a and
Jeffery L.
Yarger
*a
aDepartment of Chemistry and Biochemistry, Arizona State University, Tempe, Arizona 85287-1604, USA. E-mail: jyarger@gmail.com
bDepartment of Biology, Santa Clara University, Santa Clara, CA, USA
First published on 29th August 2014
Abstract
Embiopterans produce silken galleries and sheets using exceptionally fine silk fibers in which they live and breed. In this study, we use electron microscopy (EM), Fourier-transform infrared (FT-IR) spectroscopy, wide angle X-ray diffraction (WAXD) and solid-state nuclear magnetic resonance (ssNMR) techniques to elucidate the molecular level protein structure of webspinner (embiid) silks. Silks from two species Antipaluria urichi and Aposthonia ceylonica are studied in this work. Electron microscopy images show that the fibers are about 90–100 nm in diameter, making webspinner silks among the finest of all known animal silks. Structural studies reveal that the silk protein core is dominated by β-sheet structures, and that the protein core is coated with a hydrophobic alkane-rich surface coating. FTIR spectra of native embiid silk shows characteristic alkane CH2 stretchings near 2800–2900 cm−1, which decrease approximately 50% after washing the silk with 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CHCl3
1 CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH. Furthermore, 13C ssNMR data shows a significant CH2 resonance that is strongly affected by the presence of water, supporting the idea that the silk fibers are coated with a hydrocarbon-rich layer. Such a layer is likely used to protect the colonies from rain. FTIR data also suggests that embiid silks are dominated by β-sheet secondary structures similar to spider and silkworm silk fibers. NMR data confirms the presence of β-sheet nanostructures dominated by serine-rich repetitive regions. A deconvolution of the serine Cβ NMR resonance reveals that approximately 70% of all seryl residues exist in a β-sheet structure. This is consistent with WAXD results that suggest webspinner silks are 70% crystalline, which is the highest crystalline fraction reported for any animal silks. The work presented here provides a molecular level structural picture of silk fibers produced by webspinners.
MeOH. Furthermore, 13C ssNMR data shows a significant CH2 resonance that is strongly affected by the presence of water, supporting the idea that the silk fibers are coated with a hydrocarbon-rich layer. Such a layer is likely used to protect the colonies from rain. FTIR data also suggests that embiid silks are dominated by β-sheet secondary structures similar to spider and silkworm silk fibers. NMR data confirms the presence of β-sheet nanostructures dominated by serine-rich repetitive regions. A deconvolution of the serine Cβ NMR resonance reveals that approximately 70% of all seryl residues exist in a β-sheet structure. This is consistent with WAXD results that suggest webspinner silks are 70% crystalline, which is the highest crystalline fraction reported for any animal silks. The work presented here provides a molecular level structural picture of silk fibers produced by webspinners.
1 Introduction
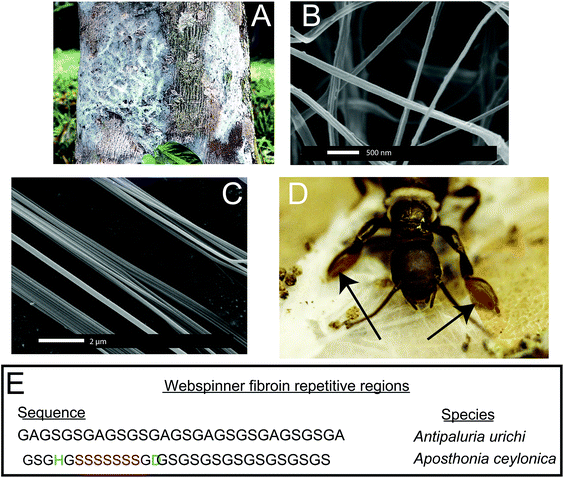
Embioptera (often called webspinners or embiids) produce silken galleries and sheets using exceptionally fine silk fibers in which they live and breed.1,2 Embiopterans are unusual insects because they are soft-bodied and flexible even as adults, and to a great extent, rely on silk instead of a tough exoskeleton for protection. Juvenile-form adult females, maternal care, and a colonial life that is defined by shared silk characterize this taxonomic order of insects. They spin silk produced in glands in the basal segment of their front feet (Fig. 1D).1 They step around the head and body as silk issues forth from multiple silk ejectors (for examples of spin step kinematics see Edgerly et al., 2012).3 Immatures, as well as adults, spin silk into domiciles that serve as retreats and egg chambers and into covered route-ways that lead to food. The two focal species in this study, Antipaluria urichi (Family Clothodidae) and Aposthonia ceylonica (Family Oligotomidae), are arboreal and tropical, living on tree bark where they graze on epiphytic algae and lichens within the relative safety of the silk (Fig. 1A). Arboreal species share silk, affording them protection from predators, especially ants, which typically walk over silk apparently without recognizing that valuable prey lay beneath.Webspinner silks are among the thinnest of all known silk-based biopolymers, but previous reports of fiber diameters are conflicting. Okada et al.4 found from SEM images that silks from the Australian webspinner Aposthonia gurneyi have fibers with a mean diameter of only 65 nm, while polarized optical microscopy data by Collin et al.5,6 found fiber diameters in the range of 500 nm for multiple embiopteran species. This discrepancy is curious, and is addressed in this work.
While the embiopteran taxonomy and behavior have been well studied,1–3,7–9 very little is known about the molecular-level structure of their silks. Significant insight was gained through cDNA sequencing of the primary silk protein sequence from multiple embiopteran species including An. urichi and Ap. ceylonica, which are the subjects of this study.6,10 Representations of the full (approximately 70 kDa) primary protein sequence for both species are seen in Fig. 1E. Like other silk-based biopolymers,11–13 webspinner silks are composed of highly repetitive protein sequences in which glycine, serine, and alanine are heavily represented.4,6,10 Although there exists very little experimental data characterizing webspinner silks, the obvious similarities between the embiopteran protein sequence and other well-characterized fibroin biopolymers allows us to make predictions. Silkworm and spider silks are also composed of highly repetitive proteins that contain runs of poly(GA) and poly(A).14,15 It is well known that these repeat motifs form water-inaccessable antiparallel β-sheet structures that are aligned along the fiber axis, and these rigid, nanocrystalline β-sheets are thought to be responsible for the impressive mechanical strengths of both spider and silkworm silks.16–18 When one looks at the cDNA sequences for many embiid silks,10 similarities are observed but alanine appears to have been mostly replaced by serine. Instead of runs of poly(GA), embiopteran primary protein sequences are dominated by runs of poly(GS) or GAGSGS repeats. Instead of runs of poly(A), some webspinner silks contain runs of poly(S). Serine has a higher propensity to adopt a β-sheet environment than alanine because of its ability to form side chain intra-sheet hydrogen bonds,19–21 therefore similar nanocrystalline β-sheet structures are fully expected from poly(GS), GAGSGS and poly(S) repeats found in embiopteran silks. Indeed, previous Fourier transform infrared spectroscopy studies on embiopteran silks have suggested that the fibers are dominated by β-sheet secondary structures within the protein core, presumably arising from said repetitive protein motifs.4–6,10
These β-sheet structures within webspinner silks are potentially nanocrystalline and well aligned with respect to the fiber axis, similarly to other well-characterized silks. The percentage of the total fiber content that is nanocrystalline varies significantly among animal silks. Studies on spider dragline silk fibers show that the crystallinity content ranges from about 30% (Nephila clavipes) to 40% (Latrodectus hesperus),23,24 while fibers from the domesticated silkworm are typically higher, in the range of 40–60% crystallinity.25,26 The crystalline fractions arise mainly from repeated sequences, thus the primary protein sequence for webspinner silks should yield predictive power; protein secondary structure within spider dragline silk fibers has been shown to correlate quantitatively with silk primary protein sequences.24,27 With the exception of short C-terminal domains, the protein sequences of many embiopteran silks are virtually entirely composed of repetitive motifs, which likely adopt a β-sheet structure. For example, if all of the repeat domains within embiopteran silk from the species An. urichi adopt a β-sheet nanocrystalline structure, then the percent crystallinity may be upwards of 90%. If true, embiopteran silks are substantially higher in nanocrystalline content than other well characterized silk biopolymers. To the best of our knowledge, no attempt has been made to measure the nanocrystallite size, orientation or crystalline fraction of any webspinner silks, which we address in this work through Wide Angle X-ray Diffraction (WAXD) techniques.
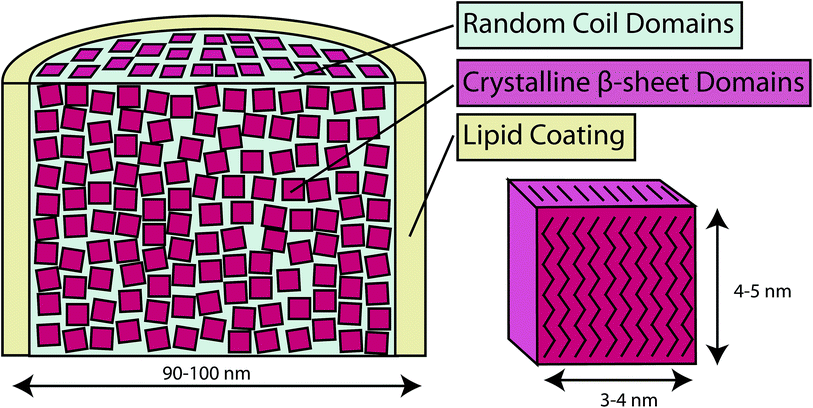
We are interested in better understanding the hierarchical structure of webspinner silks. Considering the many similarities in primary protein composition, it would not be surprising if the hierarchical structure of webspinner silk is similar to that of other well-characterized silks. The current skin-core model for spider dragline silks, for example, is a fibrous protein core that is encapsulated by a thin protective lipid-like coating.28 As discussed, the protein core of spider dragline silk is composed of nanocrystalline β-sheet structures with dimensions of a few nanometers aligned along the fiber axis, separated by loosely-organized sheet, helical or randomly-oriented domains.22,29–31 This protein core is surrounded by a thin protective coating that is rich in long chain lipids and alkanes.32–34 Very little work has been performed characterizing this outer protective coating on spider silks, although Schulz et al.34 conducted a thorough GC-MS analysis of the lipid and hydrocarbon content in silk from the orb weaver spider Nephila clavipes, revealing the presence of mostly long chain alkanes and methoxyalkanes of chain length between C28 and C34. The role of the lipid or alkane-rich surface coating is unclear, but it likely serves to both waterproof the silk fibers and maintain moisture within the silk core.32 Based on observations in the field and on laboratory colonies, embiopteran silken galleries are remarkably water-repellant, thus it is feasible that along with a β-sheet dominated protein core webspinner silk fibers possess a similar lipid or alkane-rich surface coating that serves a waterproofing purpose. Water-repellency would appear an especially adaptive trait for An. urichi, a tropical rainforest species from Trinidad and Tobago. Their colonies live on tree bark where they feed on epiphytic algae and lichens from within the protective covering of their silk. Rainfall is heavy, often torrential, and yet water appears to flow over the surface of the silk, leaving the insects dry beneath. Ap. ceylonica species are subtropical and tropical as well and are likely exposed to similar environmental conditions as the Trinidadian species. The availability of these two species in laboratory cultures and of published work on their silk provided us with an opportunity to further investigate the functional and structural aspects of embiopteran silk.
Interestingly, a few other silk-producing insects are known to contain a significant lipid content.35,36 It is often challenging to characterize a surface coating on spider silks with NMR techniques because this layer is thin relative to the overall fiber size and therefore hard to detect. The exceptionally fine fiber diameters of embiid silks provides us with a unique opportunity to observe and characterize any surface coating in addition to the protein core because of the higher surface area to volume ratio. In this present study, we use SEM, TEM, FT-IR, WAXD and solid-state NMR techniques to help elucidate both molecular-level protein structure within the silk fiber core, and to interrogate the alkane-rich surface coating surrounding the silk fibers.
2 Materials and methods
2.1 Silk collection
Cultures of approximately 50 An. urichi and Ap. ceylonica were established in separate plastic terrariums with dry oak leaves used as substrate. The insects were fed fresh romaine lettuce leaves every few days. The colony was misted with tap water every few days to maintain moisture and humidity within the domicile. The insects would spin silken sheets and tunnels, and the clean areas of these freshly spun networks were removed from the terrarium. Any visible debris (fragmented oak leaves, pebbles, dirt and waste) were removed from the silk using a pair of fine forceps under a dissecting microscope.2.2 Fourier transformed infrared spectroscopy
FT-IR analyses were performed using a Thermo Nicolet 6700 spectrometer equipped with an attenuated total reflectance (Smart Orbit ATR) accessory. For each sample, silk bundles were pressed onto a diamond window of the ATR attachment, and 64 scan averages were collected after 128 background scans.2.3 Scanning electron microscopy
Cleaned webspinner silks were secured to conductive carbon tape, then were gold coated using a Denton vacuum sputter coater for 3 minutes at a deposition rate of 5 nm min−1. The SEM was performed using a XL30 Environmental SEM-FEG built by FEI. The secondary electron (SE) detector was used for imaging. Images were collected under a vacuum pressure of less than 9 × 10−5 mbar and with a beam current of 5 kV. SEM images were processed using Gwyddion version 2.31 to measure fiber diameters from multiple images.372.4 Transmission electron microscopy
Cleaned webspinner silk samples were immersed in 0.5% aqueous uranyl acetate and incubated 30 min at room temperature. An untreated control group was run in parallel and incubated in deionized water only. Both groups were washed with 3 consecutive changes of deionized water and allowed to air-dry overnight. Samples were then incubated in 3 consecutive changes of Spurrs epoxy resin over several hours, flat-embedded on Teflon-coated glass slides, and polymerized at 60 °C for 24 h. Segments containing multiple fibers were cut from the thin resin layer and re-embedded in flat polymer molds to obtain the desired cross-sectional orientation for microtomy. Sections were cut at 70 nm thickness with a diamond knife on a Leica Ultracut-R microtome. Images were generated on a Philips CM12 TEM operated at 80 kV and captured with a Gatan model 791 CCD camera. TEM images were processed using Gwyddion version 2.31 to measure fiber diameters.372.5 Wide angle X-ray diffraction
Embiid silk fibers were aligned parallel across a cardboard washer using a small amount of super glue at either end. Wide-angle X-ray diffraction measurements were collected at the Advanced Photon Source located at Argonne National Laboratory, Argonne IL, USA on the BioCars 14BM-C beamline which has X-ray wavelength of 0.9787 Å (12.668 keV) and a beam size of 130 × 340 μm (FWHM). The samples were mounted on a goniometer 300 mm from the ADSC Quantum-315 9-panel CCD array detector with the fibers offset a small angle from the beamstop to better view the meridian reflections. The exposure time was 60 seconds for each of ten images averaged and 5 background images were taken with the same parameters to remove air scattering. Images were then processed using Fit2D X-ray processing software and calibrated with cerium dioxide (CeO2).2.6 Peptide preparation
A peptide model, GSGHGSSSSSSSGDGSGSGSGSGSGSGSGA, was synthesized using Fmoc-chemistry to mimic webspinner silk of the species Ap. ceylonica. A fully automated microwave-assisted peptide synthesizer (Liberty 1 by CEM) was used to prepare the peptide on the 0.1 mmol scale. The crude peptide was cleaved from the resin (preloaded Alanine-Wang resin from AAPPTEC) for 4 hours using a standard cleavage cocktail (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 TFA
2.5 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O). The crude peptide was precipitated out of TFA using cold diethyl ether, and after several ether washes, the crude peptide was dried, dissolved in 6 M LiBr, and dialyzed against water at 4 °C for 4 days. Any water-insoluble peptide was centrifuged out, and solubilized peptide was recovered by lyophilization. This lyophilized peptide represents Ap. ceylonica silks in a random-coil structure. Some of this peptide was dissolved in formic acid and carefully pipetted onto a clean glass surface. The peptide crystallized into a β-sheet structure upon slow evaporation of formic acid. Peptides were then characterized using NMR spectroscopy.
H2O). The crude peptide was precipitated out of TFA using cold diethyl ether, and after several ether washes, the crude peptide was dried, dissolved in 6 M LiBr, and dialyzed against water at 4 °C for 4 days. Any water-insoluble peptide was centrifuged out, and solubilized peptide was recovered by lyophilization. This lyophilized peptide represents Ap. ceylonica silks in a random-coil structure. Some of this peptide was dissolved in formic acid and carefully pipetted onto a clean glass surface. The peptide crystallized into a β-sheet structure upon slow evaporation of formic acid. Peptides were then characterized using NMR spectroscopy.
2.7 Solid-state NMR
13C solid-state NMR experiments were conducted on a 400 MHz Varian wide-bore instrument equipped with a 1.6 mm triple resonance MAS probe. Samples were spun at the magic angle at 30 kHz. 13C chemical shifts were referenced externally to TMS by setting the downfield adamantane resonance to 38.56 ppm. 1H → 13C Cross Polarization under Magic Angle Spinning (CP-MAS) conditions were optimized using U-[13C, 15N] glycine. For CP experiments, typical experimental conditions used were an initial 2 μs proton π/2 pulse, a 1 ms ramped (∼15%) spin-lock pulse at a maximum of 150 kHz on the 1H channel, and a square contact pulse set to the −1 spinning side band of the Hartmann–Hahn profile on the 13C channel. A 100 kHz spectral width was used with 20 ms acquisition time, a 5 second relaxation delay, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240 scan averages. Direct Detection under Magic Angle Spinning (DD-MAS) experiments on hydrated embiid silk samples used a 2.5 μs π/2 pulse, 20 ms acquisition time, 100 kHz spectral width, a 1 second relaxation delay, and 40
240 scan averages. Direct Detection under Magic Angle Spinning (DD-MAS) experiments on hydrated embiid silk samples used a 2.5 μs π/2 pulse, 20 ms acquisition time, 100 kHz spectral width, a 1 second relaxation delay, and 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960 scan averages. All CP and DD-MAS experiments were collected using 150 kHz TPPM38 proton decoupling during acquisition. To collect data on hydrated samples, silks were first soaked in DI water for at least 30 minutes then blotted with a Kim-Wipe to remove excess water.
960 scan averages. All CP and DD-MAS experiments were collected using 150 kHz TPPM38 proton decoupling during acquisition. To collect data on hydrated samples, silks were first soaked in DI water for at least 30 minutes then blotted with a Kim-Wipe to remove excess water.
3 Results and discussion
3.1 Fiber diameters
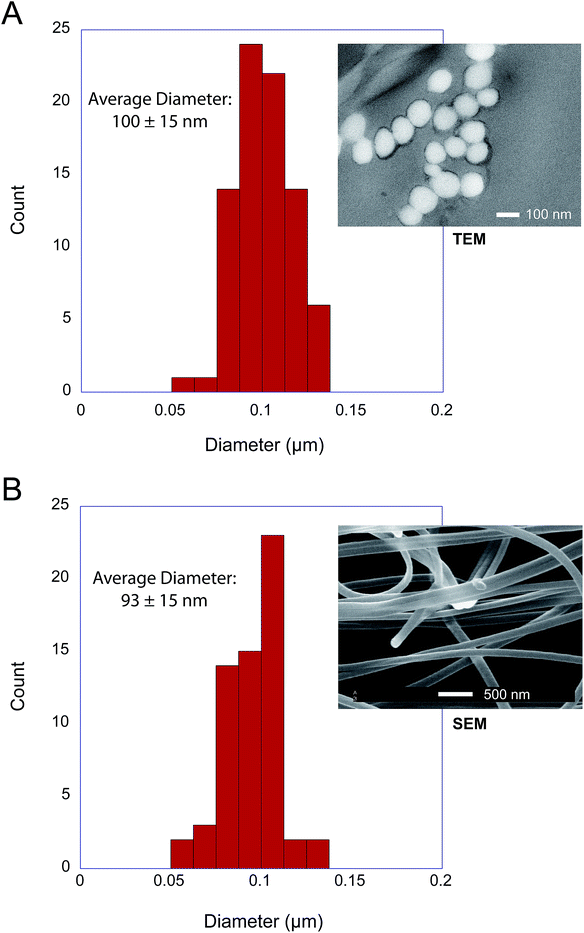
Silk from the embiopteran species Antipaluria urichi and Aposthonia ceylonica were studied using SEM, TEM, FT-IR, WAXD and NMR spectroscopy to characterize the molecular-level protein structure as well as a hydrophobic surface coating rich in long-chain lipids and alkanes. Fig. 1 shows both optical and SEM images of insects and silk produced from An. urichi. The insects produce silk out of their tarsal organs, or forelimbs, creating very thin sheets of silk protecting the colonies. An example of a silk in a natural, arboreal setting can be seen in Fig. 1A. Fiber diameters from An. urichi were determined using SEM and TEM microscopy. Previous work by Collin et al.5,6 reported fiber diameters in the range of 500–800 nm. However, the authors used polarized light microscopy techniques and thus could not resolve fibers below the optical resolution limit. Fig. 1C shows how one could easily be fooled; it is likely that the authors were observing bundles of webspinner silks and were unable to resolve fine detail. Fig. 2 shows a histogram of An. urichi fiber diameter measurements made from 68 isolated fibers over multiple SEM images and 82 fibers from TEM images. The fibers for SEM imaging had been coated with a layer of gold approximately 15 nm thick, thus 30 nm was subtracted from each edge to edge measurement. The average size was 93 ± 15 nm (one standard deviation), which is more consistent with the 65 nm fibers reported for a different webspinner species.4 For TEM imaging, fiber bundles were stained by submerging in an aqueous solution containing 0.5% uranyl acetate for 30 minutes prior to resin embedment. The average fiber diameter over 82 measurements from 3 separate TEM images was 100 ± 15 nm. This is slightly larger than the 93 nm average result from SEM images, but we note that these fibers were soaked in an aqueous-based uranyl acetate stain prior to resin embedment and thus we are potentially observing a slight swelling of the fibers due to water absorption. This observation brings doubt into the validity of previous mechanical testing results on embiid silks. Webspinner silks show elasticity similar to spider silks (15–40% extensibility), but silk strengths were reported at only about 150 MPa.5,6 If correct, this is many times weaker than spider dragline fibers and silkworm silk. As a silk used primarily for structural and protective purposes and not for absorbing impact, it would be surprising if webspinner fibers possess similar gigapascal-level strengths as spider dragline fibers. Nevertheless, embiopteran silk galleries must be strong enough to both deter predators, which often walk on top of the silk.To the best of our knowledge, the only available mechanical data obtained on silk produced by webspinner insects is unreliable due to improper fiber diameter measurements. Therefore in an attempt to better estimate embiid fiber tensile properties, we collected tensile stress–strain curves on silk bundles prepared for An. urichi. Samples were prepared by carefully brushing an E-shaped cardboard card across the tarsus of adult female insects of An. urichi. Fibers were superglued to each of the three anchor points on the E. Stress–strain curves were obtained by stretching one side of the E-shaped card at a rate of 1% per second, while the other unstretched side was analyzed using SEM to approximately obtain the number of fibers present. Additional experimental details and results are included as ESI.† Results suggest that webspinner silks are significantly stronger than previously thought; we observed an average of 500 MPa mean ultimate stress and about 30% extensibility over 14 measurements. Due to the small fiber diameters and extreme difficulty in obtaining consistent samples, this result should only be interpreted as a rough estimate.
3.2 Infrared spectroscopy: surface coating and protein core
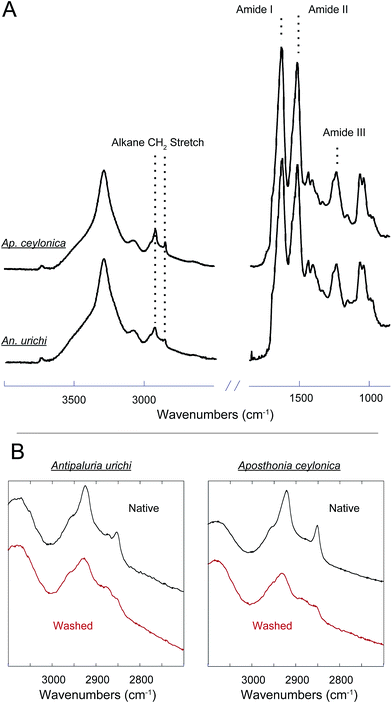
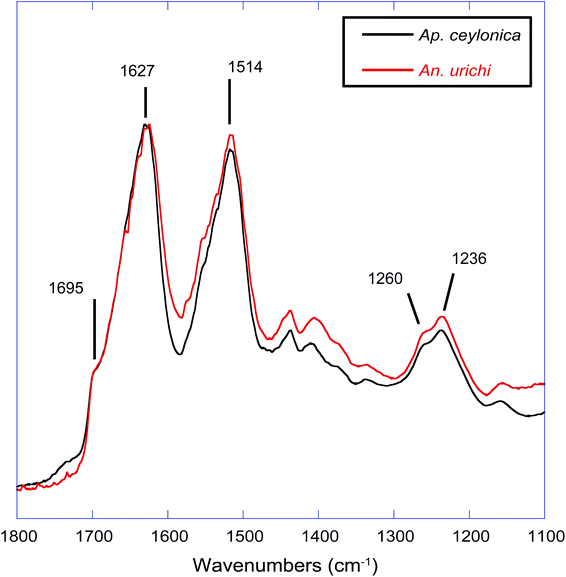
An additional key feature of embiopteran silken galleries is the ability to prevent water from penetrating into the silk networks. While composed primarily of protein, their silk is exceptionally hydrophobic; silken sheets protecting colonies do not appear to allow water to penetrate into the domiciles. If the surface of the silks were composed of glycine, alanine and serine-rich protein, one might expect water to easily penetrate a thin layer of silk. The hydrophobic nature of the silk suggests some form of lipid or alkane-rich surface coating similar to the water repelling layer found on spider silks.28,32,33 Our results provide evidence for such a surface coating. FT-IR data and NMR data both show the presence of alkanes even after washing the silk with the detergent sodium dodecyl sulfate (SDS), indicating that the surface layer must be significantly longer than C12 as to not be removed by SDS. Indeed, initial GC-MS analysis of a CHCl3 : MeOH extraction from An. urichi silk shows the presence of straight and branched-chain alkanes of at least length C34, and likely even longer (ESI†). FT-IR spectra seen in Fig. 3 on native webspinner silks show characteristic alkane CH2 symmetric and asymmetric stretch absorbances at 2850 and 2920 cm−1, respectively.39 We attribute this to alkane CH2 absorbance from the hydrocarbon-rich surface coating on the silk. Previous FT-IR studies on embiopteran silks also show a large CH2 absorbance, but it was not assigned.4 The native silk samples were then treated with 2 : 1 mixture of CHCl3 : MeOH overnight in attempt to remove any surface lipid or alkane-rich layer, and FT-IR analysis was then repeated on the treated silk samples. The amide I and amide III bands did not change suggesting that the protein structure remained unaffected (ESI†), but the alkane CH2 stretchings dropped approximately 50% in intensity after extraction (Fig. 3B). This result clearly shows the presence of a lipid or alkane-rich surface coating. It also becomes clear that the surface coating is not covalently attached to the protein core, so perhaps this surface layer is co-secreted along with the protein fiber.The protein core structure was also analyzed using infrared spectroscopy. FT-IR is an extremely common technique used to study protein secondary structure of silk-based biopolymers; some examples include silks produced from silkworm,40–43 spiders,42,44 caddisfly larvae,45 silverfish,46 lacewing,36 and webspinners.4,5 The amide I, II and amide III absorbances are commonly used to infer protein secondary structure. There seems to be a general consensus in the literature regarding the amide I band, but significant debate in assigning secondary structures to components of the amide III band. For example, the amide III β-sheet and random coil absorbances were assigned at 1263 and 1230 cm−1 respectively for silkworm silk from Bombyx mori,40 but are assigned to 1222 and 1242 cm−1 for silkworm silk from An. pernyi. It is possible that the inversion of peak location is a result of different repetitive motifs; Bombyx mori silk is dominated by poly(GA) and GAGAGS units while An. pernyi contains runs of poly(A) similar to spider silks. In support, careful secondary structure assignments to the amide III band for poly(A)-rich spider silks agree well with An. pernyi but not with Bombyx mori.42 Thus the primary protein sequence has a major impact on FT-IR absorption peak positions, as does fiber strain and overall secondary structure.47 Considering significant variation in peak position, width, and shape, we think that a quantitative deconvolution of FT-IR absorbance bands will not give convincing results. Qualitative and especially comparative analysis of IR absorbance profiles, however, can be extremely powerful. Fig. 4 shows that the FT-IR absorbance profile of An. urichi is virtually identical to that of Ap. ceylonica. The strong amide I absorbance maximum at 1627 wavenumbers is identical to reports from Okada et al.,4 on webspinner silk from the congener Ap. gurneyi, which is similarly dominated by poly(GS) and poly(S) motifs. For comparison, the amide I band for β-sheet-rich silkworm silk is found at 1615 cm−1 (ESI†). The shift towards higher frequency absorbance for webspinner silk relative to silkworm silk could be attributed to a larger random coil component, as is often done in the literature through peak deconvolution. While this is a possibility, we think it is more likely that the shift in frequency is a result of differing primary protein sequences rather than a lower β-sheet fraction, especially when considering the high expected crystalline fraction for webspinner silks. Amide I absorbances from random-coil structures are typically found near 1650 cm−1 while β-sheet structures show absorbances at lower frequencies (1620 cm−1).48 Additionally, the shoulder absorbance at 1695 cm−1 is regularly assigned to β-sheet protein structures,40,48 thus it is quite clear that like silkworm silks, webspinner silk fibers are dominated by β-sheet structures, and are remarkably similar across species.
3.3 Wide angle X-ray diffraction
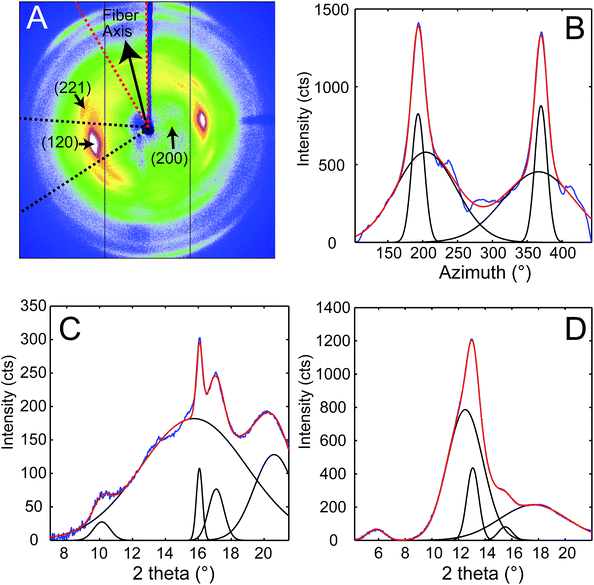
Characterization of webspinner silk from An. urichi by wide-angle X-ray diffraction (WAXD) indicates an amorphous fraction with diffuse scattering and a crystalline fraction comprised of nanocrystalline β-sheet structures. The diffraction pattern observed for axially-aligned webspinner silk from An. urichi shown in Fig. 5A closely resembles our previously published spider silk patterns, confirming the presence of regular nanocrystalline β-sheet structures within the silk protein core.23,49 Careful analysis of the WAXD profile gives crystallite size, orientation of the nanocrystallite with respect to the fiber axis, and the overall percent crystallinity of the fiber. The 12 highest intensity unique reflections were identified and assigned to an orthorhombic unit cell of dimensions a = 6.6 Å, b = 9.6 Å, c = 19.2 Å. Each reflection was fit as an individual component in d spacing using an iterative Gaussian fitting protocol until it converged. The radial broadening of the crystalline reflections, as demonstrated by the full width at half max (FWHM) in 2θ space (Fig. 5C and D), was used to calculate the average crystallite size along the a, b, and, c axes using Scherrers formula. This resulted in crystallite dimensions of 3, 4, and 5 nm in the a, b, and c axes, respectively.25 Azimuthal broadening of the crystalline reflections is indicative of variance in alignment of the crystalline fraction with respect to the fiber axis. Hermans orientation factor of the crystalline fraction, fc, was calculated to be 0.93, from the FWHM of radial integration at 4.3 Å−1 where fc = (3(cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Φ)−1)/2 and Φ is the angle between the c axis of the nanocrystallite and the fiber axis. Difficulty manually aligning the silk fibers during sample preparation leads to fiber misalignment, which means this is a low estimate of the actual nanocrystallite orientation within the fiber. The overall crystalline fraction can be estimated as a fraction of the area represented by radial integration 15° on either side of the equator for the equatorial (200) and (120) crystalline peaks relative to the area represented by the integrated intensity of the full diffraction pattern resulting in 69% crystallinity. This high crystallinity content is likely due to the high fraction of repetitive motifs and significant serine content, both of which correlate to increased β-sheet content in the silk protein core.
Φ)−1)/2 and Φ is the angle between the c axis of the nanocrystallite and the fiber axis. Difficulty manually aligning the silk fibers during sample preparation leads to fiber misalignment, which means this is a low estimate of the actual nanocrystallite orientation within the fiber. The overall crystalline fraction can be estimated as a fraction of the area represented by radial integration 15° on either side of the equator for the equatorial (200) and (120) crystalline peaks relative to the area represented by the integrated intensity of the full diffraction pattern resulting in 69% crystallinity. This high crystallinity content is likely due to the high fraction of repetitive motifs and significant serine content, both of which correlate to increased β-sheet content in the silk protein core.
3.4 Solid-state NMR
To get a more complete understanding of both the silk protein core and the encapsulating surface coating, we used solid-state NMR techniques on both native and CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
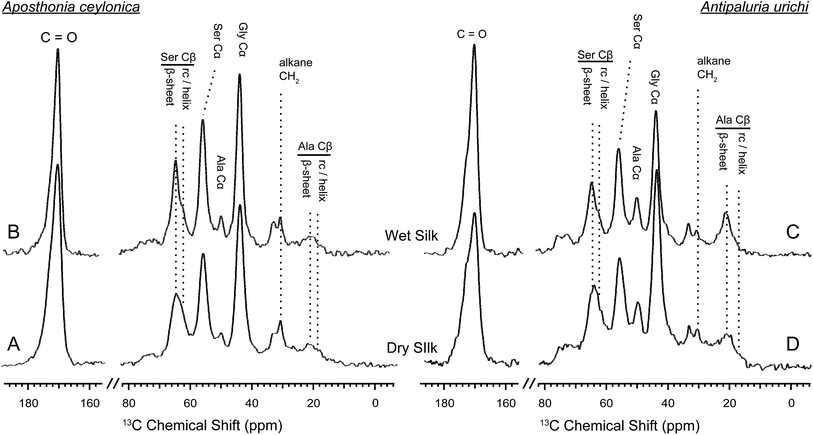
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH-washed silks in their dry and hydrated states. Solid-state NMR is a very powerful tool for characterizing biopolymers; it has been widely utilized to elucidate molecular-level environments of repeat motifs such as poly(GA) and poly(A) in silkworm and spider silks, respectively. For embiid silks from both species discussed in this work, glycine and serine are the most dominant amino acids represented at approximately 45% and 36%, respectively.10 The 13C chemical shift of glycine Cα is not very sensitive to secondary structure, however both the alanine and serine Cα and especially Cβ chemical shifts are influenced dramatically by local conformation,50,5113C chemical shifts have been used extensively to identify secondary structures within silk-based biopolymers, and can be applied here to elucidate molecular level environments within embiid silks. Cleaned, native silk from Ap. ceylonica and An. urichi were studied with 13C solid-state NMR spectroscopy. 13C resonances for the three most abundant amino acids are assigned, and secondary structures are indicated (Fig. 6, 7). We are limited in the amount of information we can extract from 13C data on naturally-abundant samples, but we can still draw many strong conclusions. For one, serine exists predominantly in β-sheet structures based on the 13C chemical shifts observed for serine Cβ (Fig. 6). As indicated in Fig. 6, the serine Cβ resonances in the CP-MAS spectra lie at 64.5 ppm, a shift consistent with serine in a β-sheet environment. Serine from GAGSGS and poly(S) repeat motifs are most likely responsible for this observation. We also can see a minor shoulder at 62 ppm that is assigned to serine containing regions in a randomly-oriented environment.
MeOH-washed silks in their dry and hydrated states. Solid-state NMR is a very powerful tool for characterizing biopolymers; it has been widely utilized to elucidate molecular-level environments of repeat motifs such as poly(GA) and poly(A) in silkworm and spider silks, respectively. For embiid silks from both species discussed in this work, glycine and serine are the most dominant amino acids represented at approximately 45% and 36%, respectively.10 The 13C chemical shift of glycine Cα is not very sensitive to secondary structure, however both the alanine and serine Cα and especially Cβ chemical shifts are influenced dramatically by local conformation,50,5113C chemical shifts have been used extensively to identify secondary structures within silk-based biopolymers, and can be applied here to elucidate molecular level environments within embiid silks. Cleaned, native silk from Ap. ceylonica and An. urichi were studied with 13C solid-state NMR spectroscopy. 13C resonances for the three most abundant amino acids are assigned, and secondary structures are indicated (Fig. 6, 7). We are limited in the amount of information we can extract from 13C data on naturally-abundant samples, but we can still draw many strong conclusions. For one, serine exists predominantly in β-sheet structures based on the 13C chemical shifts observed for serine Cβ (Fig. 6). As indicated in Fig. 6, the serine Cβ resonances in the CP-MAS spectra lie at 64.5 ppm, a shift consistent with serine in a β-sheet environment. Serine from GAGSGS and poly(S) repeat motifs are most likely responsible for this observation. We also can see a minor shoulder at 62 ppm that is assigned to serine containing regions in a randomly-oriented environment.
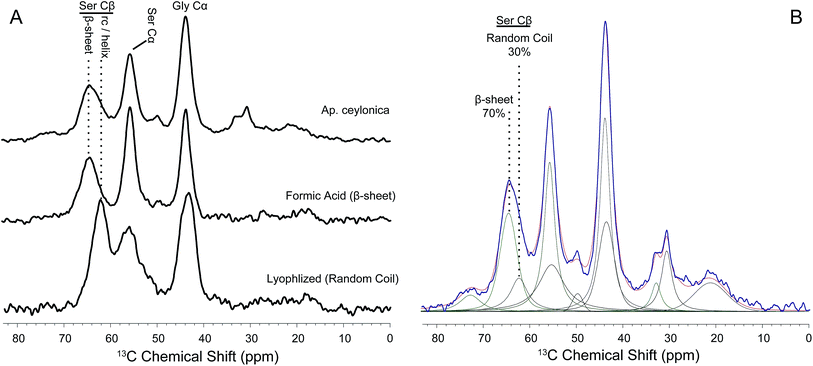
Further confirmation of the secondary structural assignments where made by comparing 1H → 13C CP-MAS NMR data of peptides mimicking webspinner repetitive motifs that were either trapped in a random-coil confirmation or crystallized into a β-sheet structure from formic acid (Fig. 8). To mimic webspinner silk in a random-coil structure, a 30 AA peptide representing the protein sequence of Ap. ceylonica silk (illustrated in Fig. 1E) was synthesized using solid-phase peptide synthesis, dissolved in 6 M LiBr, dialyzed against DI water for 4 days, and then lyophilized. The peptide was then dissolved in formic acid and slowly dried, inducing a β-sheet structure. This method is commonly used to capture silkworm model peptides in both random-coil and β-sheet conformations.52,531H → 13C CP-MAS data was collected on both peptides (8A). Using DMFIT software,54 the spectra for random-coil and β-sheet model peptides were independently fit to extract precise 13C chemical shifts, peak widths, and % Gaussian vs. % Lorentzian line shapes. This information was then used to deconvolute NMR spectra for native silks to approximate β-sheet content; only peak amplitudes were allowed to vary when fitting native webspinner silk data. Deconvolution of NMR data obtained for Ap. ceylonica (8B) and An. urichi (data not shown) silks indicate that serine Cβ, the residue most isolated and most sensitive to secondary structure, is approximately 70% β-sheet and 30% random-coil for both species. This estimation is consistent with both our FT-IR and our XRD data in that FT-IR absorbance profiles for the two species suggest that the silk protein structures are virtually identical and are dominated by β-sheet structures, and that analysis of XRD results reveal that An. urichi silk is 70% crystalline. The alanine Cβ resonance shows a similar story in both silks, although it is more clear for An. urichi silk fibers where alanine is better represented (6C and 6D). Alanine, found almost exclusively in GAGSGS repeat motifs from An. urichi silk, clearly exists predominantly in β-sheet structures.
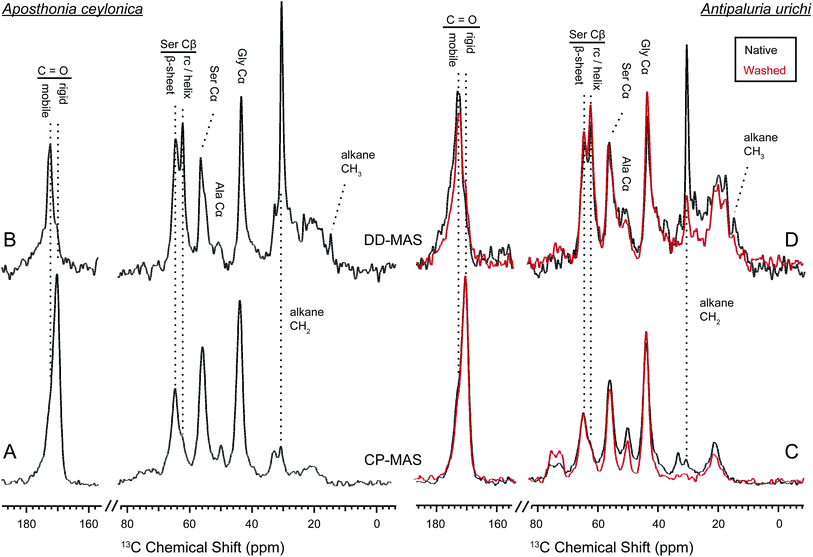
The contrast between CP and DD-MAS spectra in Fig. 7 is very useful to highlight rigid versus mobile regions of hydrated silks.55–58 Domains of the silk that are affected by water will typically have shortened 13C T1 and longer T2 relaxation times due to increased molecular motion, thus are easily seen as sharp peaks in the direct spectra. The DD-MAS experiment uses a fast (1 second) recycle delay so that 13C resonances from rigid regions of the silk with long T1 relaxation properties become saturated and therefore are not observed. While the majority of serine adopts a β-sheet structure within the silks, a new sharp resonance emerges in the direct spectra from both species at 62.2 ppm. This peak corresponds to serine Cβ in a random coil or loose helical environment. Serine is found almost exclusively in poly(GS), poly(GAGSGS) or poly(S) repetitive motifs in both silks, therefore not all seryl residues from these repeats exist in a β-sheet structure. A native silk gallery will naturally repel water due to its hydrophobic coating, but in this case we encouraged hydration of the fibers by completely submerging the silk samples in water. Perhaps when the silk becomes forcibly hydrated, poly(GS) and GAGSGS repeats from Ap. ceylonica and An. urichi silks are stable in both conformations, or can exchange between sheet-like and randomly-oriented structures on a slow timescale. Alanine resonances are weak from Ap. ceylonica silks, but we also observe a sharp peak emerge at 17.5 ppm for alanine Cβ from An. urichi silk in the direct spectra. Similar to the random coil serine Cβ peak evident by the sharp peak at 62.2 ppm, a portion of alanine also appears to adopt a random coil environment. Alanine from An. urichi silks comes from GAGSGS repeat domains, further supporting the idea that such repeats are found in both sheet-like and random structures when hydrated. For comparison, Bombyx mori silkworm silk is dominated by similar GAGAGS repeat domains, however a parallel experiment conducted on these fibers does not result in an emerging sharp helix or random-coil resonance upon fiber wetting. It appears then that when water penetrates into and solvates webspinner silk fibers, a higher fraction of silk protein becomes mobile as compared to similar regions of silkworm silk. Webspinner silk protein is much smaller than that of silkworm (70 kDa vs. 350 kDa). Perhaps for both silkworm and webspinner silks then, the dominant protein repeat units are embedded primarily in β-sheet nanostructures, but C- and N-terminal ends are less restricted. The shorter length of the webspinner protein and therefore higher percentage of protein near the C- and N-terminal domains may account for the apparent increase in random-coil protein content after fiber wetting.
Fig. 7 also yields information on the surface coating surrounding the silk fibers. For both Ap. ceylonica and An. urichi silks, one notices that the alkane CH2 resonance is significantly pronounced in the direct spectra when the silk is wet. The alkane CH2 resonance from both silks is both sharp and strong in the direct spectra, presumably because of fast molecular motion induced by the presence of water on the silk surface. Additionally, both An. urichi and Ap. ceylonica silks show a new sharp resonance in the direct spectra at 14 ppm, which we assign to terminal or branched alkane CH3 groups on the silk surface. Similar to FT-IR results in Fig. 3, NMR data shown in Fig. 7 on Ap. ceylonica silk fibers reveal the severe reduction of alkane CH2 and CH3 signals after washing the fibers with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of CHCl3
1 mixture of CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (red). We conclude that the surface coating surrounding the silk protein core is composed of long-chain lipids or alkanes that are non-covalently attached to the protein.
MeOH (red). We conclude that the surface coating surrounding the silk protein core is composed of long-chain lipids or alkanes that are non-covalently attached to the protein.
4 Conclusions
An in depth analysis has been performed on webspinner silks from An. urichi and Ap. ceylonica, and our findings, summarized in Fig. 9, allow us to improve the current understanding of the molecular-level and hierarchical structure of embiopteran silk fibers. Our best electron microscopy results show that webspinner silks are 90–100 nm in diameter, making them among the thinnest known silk-based biopolymers. Both FT-IR and NMR results confirm what could be predicted by the repetitive primary protein sequences; the protein core is dominated by nanocrystalline β-sheet structures arising from glycine- and serine-rich repeat motifs. Wide angle X-ray diffraction results show that for An. urichi silk, and presumably other embiopteran silks, these nanocrystallites are highly ordered, are well aligned with respect to the fiber axis (low estimate: fc = 0.93), and the crystallites are approximately 3 to 5 nm in dimension. Additionally, X-ray data show that webspinner silks possess an extremely high crystalline fraction at 69%. This is consistent with solid-state NMR results, which indicate that 70% of serine residues reside in nanocrystalline β-sheet structures. The silk is naturally water-repellent, but when the silk is water saturated, some regions of the protein, possibly the N- and C-termini of the relatively small (70 kDa) protein, become mobile and randomly-oriented. NMR and FT-IR data also provide evidence for a long-chain lipid or alkane-rich surface coating on webspinner silk fibers. Characteristic alkane infrared absorbances and 13C resonances decrease 50% in intensity after treating the silk with 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CHCl3
1 CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH, suggesting that this surface coating is non-covalently adhered to the protein core. The hydrophobic nature of silken galleries appears to serve to protect the insects from being dislodged from their arboreal dwellings during heavy rain. It also protects the interior of their domiciles and their bodies from extreme wetting, which has multiple negative consequences including increased risk of suffocation and of fungal attack. During severe dry periods, the silk might also serve to limit desiccation of the insects sheltering inside. The basic hierarchical structure of webspinner silk fibers is illustrated in this study: the thin fiber core is composed of glycine and serine-rich repetitive protein motifs that primarily adopt a β-sheet nanostructure, and the protein core surrounded by a thin protective hydrophobic lipid or alkane-rich shell.
MeOH, suggesting that this surface coating is non-covalently adhered to the protein core. The hydrophobic nature of silken galleries appears to serve to protect the insects from being dislodged from their arboreal dwellings during heavy rain. It also protects the interior of their domiciles and their bodies from extreme wetting, which has multiple negative consequences including increased risk of suffocation and of fungal attack. During severe dry periods, the silk might also serve to limit desiccation of the insects sheltering inside. The basic hierarchical structure of webspinner silk fibers is illustrated in this study: the thin fiber core is composed of glycine and serine-rich repetitive protein motifs that primarily adopt a β-sheet nanostructure, and the protein core surrounded by a thin protective hydrophobic lipid or alkane-rich shell.
Acknowledgements
We gratefully acknowledge the School of Life Sciences EM Laboratory at Arizona State University for TEM imaging, and the LeRoy Eyring Center for Solid State Science at Arizona State University for SEM imaging. We also thank Dr Brian Cherry for help with NMR and Robert Henning for his help during the X-ray experiments. Use of the Argonne National Laboratory (ANL) Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357 and DOE-EPSCoR under contract number DE-SC0004791. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR007707. The Department of Defense (DOD) Air Force Office of Scientific Research (AFOSR) under award no. FA9550-14-1-0014 and the National Science Foundation (NSF), Division of Materials Research under award no. DMR-1264801 supported this work. NSF grant DEB-0515865 awarded to JSE at SCU supported collecting trips into the field and maintenance of cultures at SCU. We also thank Khaaliq Dejan for assistance in harvesting and cleaning silk. He was supported by the Faculty Student Research Assistant Program at SCU.References
- J. S. Edgerly, J. A. Davilla and N. Schoenfeld, J. Insect Behav., 2002, 15, 219–242 CrossRef.
- J. S. Edgerly, The Evolution of Social Behavior in Insects and Arachnids, Cambridge University Press, 1997, pp. 14–25 Search PubMed.
- J. S. Edgerly, S. Büsse and T. Hörnschemeyer, Zool. Anz., 2012, 297–306 CrossRef PubMed.
- S. Okada, S. Weisman, H. E. Trueman, S. T. Mudie, V. S. Haritos and T. D. Sutherland, Int. J. Biol. Macromol., 2008, 43, 271–275 CrossRef CAS PubMed.
- M. A. Collin, E. Camama, B. O. Swanson, J. S. Edgerly and C. Y. Hayashi, Biomacromolecules, 2009, 10, 2268–2274 CrossRef CAS PubMed.
- M. A. Collin, J. E. Garb, J. S. Edgerly and C. Y. Hayashi, Insect Biochem. Mol. Biol., 2009, 39, 79–82 CrossRef PubMed.
- J. S. Edgerly, A. Tadimalla and E. P. Dahlhoff, Funct. Ecol., 2005, 19, 255–262 CrossRef PubMed.
- E. S. Ross, Encyclopedia of Entomology, Springer, Netherlands, 2008, pp. 4169–4172 Search PubMed.
- K. B. Miller, C. Y. Hayashi, M. F. Whiting, G. J. Svenson and J. S. Edgerly, Syst. Entomol., 2012, 37, 550–570 CrossRef PubMed.
- M. A. Collin, J. S. Edgerly and C. Y. Hayashi, Zoology, Jena, Germany, 2011, 114, pp. 239–246 Search PubMed.
- R. V. Lewis, Chem. Rev., 2006, 106, 3762–3774 CrossRef CAS PubMed.
- G. H. Altman, F. Diaz, C. Jakuba, T. Calabro, R. L. Horan, J. Chen, H. Lu, J. Richmond and D. L. Kaplan, Biomaterials, 2003, 24, 401–416 CrossRef CAS.
- T. D. Sutherland, J. H. Young, S. Weisman, C. Y. Hayashi and D. J. Merritt, Annu. Rev. Entomol., 2010, 55, 171–188 CrossRef CAS PubMed.
- C. Z. Zhou, F. Confalonieri, N. Medina, Y. Zivanovic, C. Esnault, T. Yang, M. Jacquet, J. Janin, M. Duguet, R. Perasso and Z. G. Li, Nucleic Acids Res., 2000, 28, 2413–2419 CrossRef CAS PubMed.
- M. Xu and R. V. Lewis, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7120–7124 CrossRef CAS.
- S. Keten, Z. Xu, B. Ihle and M. J. Buehler, Nat. Mater., 2010, 9, 359–367 CrossRef CAS PubMed.
- S. Keten and M. J. Buehler, Appl. Phys. Lett., 2010, 96, 153701 CrossRef PubMed.
- C. Y. Hayashi, N. H. Shipley and R. V. Lewis, Int. J. Biol. Macromol., 1999, 24, 271–275 CrossRef CAS.
- J. Johansson, C. Nerelius, H. Willander and J. Presto, Biochem. Biophys. Res. Commun., 2010, 402, 515–518 CrossRef CAS PubMed.
- R. W. Williams, A. Chang, D. Juretić and S. Loughran, Biochim. Biophys. Acta, 1987, 916, 200–204 CrossRef CAS.
- D. L. Minor Jr and P. S. Kim, Nature, 1994, 367, 660–663 CrossRef CAS PubMed.
- D. Grubb and L. Jelinski, Macromolecules, 1997, 30, 2860–2867 CrossRef CAS.
- S. Sampath, T. Isdebski, J. E. Jenkins, J. V. Ayon, R. W. Henning, J. P. R. O. Orgel, O. Antipoa and J. L. Yarger, Soft Matter, 2012, 8, 6713–6722 RSC.
- J. E. Jenkins, S. Sampath, E. Butler, J. Kim, R. W. Henning, G. P. Holland and J. L. Yarger, Biomacromolecules, 2013, 14, 3472–3483 CrossRef CAS PubMed.
- K. Liang, Y. Gong, J. Fu, S. Yan, Y. Tan, R. Du, X. Xing, G. Mo, Z. Chen, Q. Cai, D. Sun and Z. Wu, Int. J. Biol. Macromol., 2013, 57, 99–104 CrossRef CAS PubMed.
- T. Asakura, J. Yao, T. Yamane, K. Umemura and A. S. Ulrich, J. Am. Chem. Soc., 2002, 124, 8794–8795 CrossRef CAS PubMed.
- J. E. Jenkins, M. S. Creager, R. V. Lewis, G. P. Holland and J. L. Yarger, Biomacromolecules, 2009, 11, 192–200 CrossRef PubMed.
- A. Sponner, W. Vater, S. Monajembashi, E. Unger, F. Grosse and K. Weisshart, PLoS One, 2007, 2, e998 Search PubMed.
- G. P. Holland, R. V. Lewis and J. L. Yarger, J. Am. Chem. Soc., 2004, 126, 5867–5872 CrossRef CAS PubMed.
- Z. Yang, D. Grubb and L. Jelinski, Macromolecules, 1997, 30, 8254–8261 CrossRef CAS.
- C. Riekel, C. Bränden, C. Craig, C. Ferrero, F. Heidelbach and M. Müller, Int. J. Biol. Macromol., 1999, 24, 179–186 CrossRef CAS.
- F. Vollrath, T. Holtet, H. C. Thogersen and S. Frische, Proc. R. Soc. London, Ser. B, 1996, 263, 147–151 CrossRef.
- S. Frische, A. B. Maunsbach and F. Vollrath, J. Microsc., 1998, 189, 64–70 CrossRef CAS.
- S. Schulz, Lipids, 2001, 36, 637–647 CrossRef CAS PubMed.
- E. Victoriano, D. O. Pinheiro and E. A. Gregório, Neotrop. Entomol., 2007, 36, 707–711 CrossRef PubMed.
- S. Weisman, H. E. Trueman, S. T. Mudie, J. S. Church, T. D. Sutherland and V. S. Haritos, Biomacromolecules, 2008, 9, 3065–3069 CrossRef CAS PubMed.
- D. Nečas and P. Klapetek, Cent. Eur. J. Phys., 2011, 10, 181–188 CrossRef PubMed.
- A. E. Bennett, C. M. Rienstra, M. Auger, K. V. Lakshmi and R. G. Griffin, J. Chem. Phys., 1995, 103, 6951 CrossRef CAS PubMed.
- R. N. A. H. Lewis and R. N. McElhaney, Methods in molecular biology, Clifton, N.J., 2007, vol. 400, pp. 207–226 Search PubMed.
- M. Boulet-Audet, T. Lefèvre, T. Buffeteau and M. Pézolet, Appl. Spectrosc., 2008, 62, 956–962 CrossRef CAS.
- J. Shao, J. Zheng, J. Liu and C. M. Carr, J. Appl. Polym. Sci., 2005, 96, 1999–2004 CrossRef CAS.
- S. Ling, Z. Qi, D. P. Knight, Z. Shao and X. Chen, Biomacromolecules, 2011, 12, 3344–3349 CrossRef CAS PubMed.
- S. Ling, Z. Qi, D. P. Knight, Y. Huang, L. Huang, H. Zhou, Z. Shao and X. Chen, Biomacromolecules, 2013, 14, 1885–1892 CrossRef CAS PubMed.
- P. Papadopoulos, R. Ene, I. Weidner and F. Kremer, Macromol. Rapid Commun., 2009, 30, 851–857 CrossRef CAS PubMed.
- N. N. Ashton, D. R. Roe, R. B. Weiss, T. E. Cheatham III and R. J. Stewart, Biomacromolecules, 2013, 14, 3668–3681 CrossRef CAS PubMed.
- A. A. Walker, J. S. Church, A. L. Woodhead and T. D. Sutherland, Insect Biochem. Mol. Biol., 2013, 43, 572–579 CrossRef CAS PubMed.
- P. Papadopoulos, J. Sölter and F. Kremer, Eur. Phys. J. E: Soft Matter Biol. Phys., 2007, 24, 193–199 CrossRef CAS PubMed.
- V. Syu and N. N. Kalnin, Biopolymers, 1990, 30, 1259–1271 CrossRef PubMed.
- J. O. Warwicker, J. Mol. Biol., 1960, 2, 350–362 CrossRef CAS.
- H. Saito, Magn. Reson. Chem., 1986, 24, 835–852 CrossRef CAS.
- A. Shoji, T. Ozaki, H. Saito, R. Tabeta and I. Ando, Macromolecules, 1984, 17, 1472–1479 CrossRef CAS.
- T. Asakura, K. Suita, T. Kameda, S. Afonin and A. S. Ulrich, Magn. Reson. Chem., 2004, 42, 258–266 CrossRef CAS PubMed.
- T. Asakura, R. Sugino, T. Okumura and Y. Nakazawa, Protein Sci., 2002, 11, 1873–1877 CrossRef CAS PubMed.
- D. Massiot, F. Fayon, M. Capron, I. King, S. Le Calvé, B. Alonso, J. O. Durand, B. Bujoli, Z. Gan and G. Hoatson, Magn. Reson. Chem., 2002, 40, 70–76 CrossRef CAS.
- M. S. Creager, J. E. Jenkins, L. A. Thagard-Yeaman, A. E. Brooks, J. A. Jones, R. V. Lewis, G. P. Holland and J. L. Yarger, Biomacromolecules, 2010, 11, 2039–2043 CrossRef CAS PubMed.
- G. P. Holland, J. E. Jenkins, M. S. Creager, R. V. Lewis and J. L. Yarger, Biomacromolecules, 2008, 9, 651–657 CrossRef CAS PubMed.
- Z. Yang, O. Liivak, A. Seidel, G. LaVerde, D. Zax and L. Jelinski, J. Am. Chem. Soc., 2000, 122, 9019–9025 CrossRef CAS.
- J. B. Addison, N. N. Ashton, W. S. Weber, R. J. Stewart, G. P. Holland and J. L. Yarger, Biomacromolecules, 2013, 14, 1140–1148 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra07567f |
| This journal is © The Royal Society of Chemistry 2014 |