 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescence detection of natural RNA using rationally designed “clickable” oligonucleotide probes†
Anders
Okholm
a,
Jørgen
Kjems
a and
Kira
Astakhova
*b
aInterdisciplinary Nanoscience Center (iNANO) and Department of Molecular Biology and Genetics, Aarhus University, Gustav Wiedsvej 14, DK-8000 Århus C, Denmark. E-mail: jk@mb.au.dk; Tel: +45 28992086
bNucleic Acid Center, Department of Physics, Chemistry and Pharmacy, University of Southern Denmark, DK-5230 Odense M, Denmark. E-mail: ias@sdu.dk; Tel: +45 6550 2510
First published on 24th September 2014
Abstract
Herein a reliable approach to the design of effective fluorescent probes for RNA detection is described. The fluorescence signalling of hybridization by internally positioned polyaromatic hydrocarbons and rhodamine dyes was achieved with a low fluorescence background signal, high fluorescence quantum yields at ambient and elevated temperature, high selectivity and signal specificity of the probes when binding to miR-7 and circRNA targets.
RNA targeting is a rapidly evolving field of research and clinical diagnostics which applies synthetic fluorescent probes and modern biophysical techniques to monitor desired targets in vitro and in vivo.1 Preparation of fluorescent oligonucleotides with high potential in RNA detection by a simple and rapid method, e.g. by fluorescence, is an important aspect of current nucleic acid chemistry. Recently copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) click chemistry has been successfully applied for nucleic acid labelling, allowing preparation of various oligonucleotides in high yields and purity.2,3 Furthermore, attachment of fluorophores by click chemistry to 2′-O-propargyl uridine scaffold UP was proven to yield probes with promising properties in diagnostics (Fig. 1).4 However, in order to further utilize “clickable” oligonucleotides in diverse analytical experiments, the fluorescent probes have to meet several additional requirements, such as high photo- and chemical stability, low fluorescence background signal from single stranded molecules, high binding specificity and low limit of target detection.5 Ideally, incorporation of fluorescent dyes should furthermore be simple and allow high flexibility in probe design.
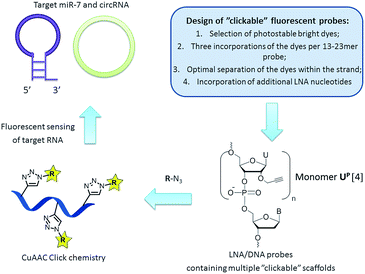 | ||
| Fig. 1 General strategy for targeting natural RNA molecules (i.e. miR-7 and circRNA) by “clickable” fluorescent probes. | ||
MicroRNAs (miRNAs) are short (∼22 nucleotides) noncoding RNA molecules that post-transcriptionally repress the expression of protein-coding genes by binding to 3′-untranslated regions of the target mRNAs.6 An increasing amount of evidence shows that miRNAs control a large number of biological processes, including cell differentiation, tumourigenesis and regulation of metabolism. A thoroughly studied miRNA is miRNA-7 (miR-7) which is highly expressed in brain neurons and also an important negative regulator of oncogenes and, hence, functioning as a tumor suppressor factor in oncogenesis.7 miR-7 was recently identified to be selectively expressed within the hypothalamus, a part of the brain that controls vital bodily functions.7
Another class of recently discovered RNA molecules with important, yet not fully studied, regulatory functions are circular RNAs (circRNAs).8,9 Interestingly, it was found that a human circRNA, antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as), is densely bound by miR-7 molecules in cells and presumably acts as a regulator of miR-7 activity.9 Clearly, in situ visualization of miR-7 and CDR1as will significantly contribute current understanding of regulatory RNA pathways and may provide further insight into the regulatory role of circRNA.6–9 In the present work we aimed at a robust approach to preparation of fluorescent probes with high binding affinity and efficient fluorescence detection of miR-7 and circRNA (Fig. 1). Herein, the design and synthesis of miR-7 and circRNA targeting oligonucleotides containing 2′-O-propargyl uridine scaffolds UP, their fluorescent labelling by CuAAC click reactions, and spectroscopic characterization of the resulting fluorescent probes in vitro are reported. The ability to design and rapidly prepare bright selective fluorescent probes for detecting any natural RNA target could strongly support research in this direction.6–9
First, we selected perylene and two rhodamine derivatives, 5-R110 and 6-ROX, as fluorophores with interesting optical properties to be internally attached to the scaffold UP within new probes (Fig. 1 and Scheme 1).4,10,11 Second, three incorporations of the dyes within 13–21mer oligonucleotides were previously demonstrated to be optimal for spectral properties of the probes.4 Therefore commercially available phosphoramidite reagent 1 was triply incorporated into the 23mer miR-7 and circRNA targeting sequences resulting in oligonucleotides ON1, ON2 (Fig. 1 and Table 1; ESI, Table S1†).6–9 Third, the “clickable” scaffolds UP within ON1, ON2 were separated by 1–2 nucleotides in order to achieve optimal fluorescence emitted from the probes upon target binding.4 As a final design aspect, additional LNA (locked nucleic acids; Scheme 1) monomers were incorporated into ON1, ON2 to enhance binding affinity and specificity of the probes to complementary targets.11 Finally, reference probes ON3, ON4 were prepared in order to evaluate influence of the modified monomers UP and M1–M3 on target binding (Table 1).
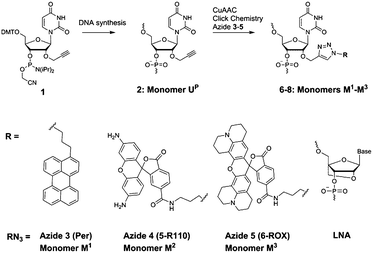 | ||
| Scheme 1 Modified monomers applied in this study; postsynthetic CuAAC click chemistry generating fluorescent miR-7 and circRNA oligonucleotides. LNA = locked nucleic acid. | ||
| # | Sequence 5′ → 3′ | Target | T m, °C vs. complementary | |
|---|---|---|---|---|
| DNA | RNA | |||
| a T m of unmodified duplexes formed by miR-7 and circRNA: 61.0 °C and 60.0 °C with complementary DNA, and 57 °C and 55.5 °C with complementary RNA, respectively; LNA monomers are marked with an L in superscript. | ||||
| ON1 | ACAACAAALATCLAC![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PAG PAG![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PC PC![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PTCCLA PTCCLA |
miR-7 | 62.0 | 66.5 |
| ON2 | GAAGACLGTGLGA![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PT PT![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PTC PTC![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) PGGALAGA PGGALAGA |
circRNA | 62.0 | 66.0 |
| ON3 | ACAACAAALATCLACTAGTCTTCCLA | miR-7 | 67.0 | 66.0 |
| ON4 | GAAGACLGTGLGATTTTCTGGALAGA | circRNA | 70.0 | 72.0 |
| P1 | ACAACAAALATCLAC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1AG 1AG![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1C 1C![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1TCCLA 1TCCLA |
miR-7 | 68.0 | 68.0 |
| P2 | GAAGACLGTGLGA![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1T 1T![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1TC 1TC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 1GGALAGA 1GGALAGA |
circRNA | 68.0 | 64.0 |
| P3 | ACAACAAALATCLAC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2AG 2AG![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2C 2C![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2TCCLA 2TCCLA |
miR-7 | 57.0 | 62.0 |
| P4 | GAAGACLGTGLGA![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2T 2T![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2TC 2TC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 2GGALAGA 2GGALAGA |
circRNA | 57.0 | 61.0 |
| P5 | ACAACAAALATCLAC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3AG 3AG![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3C 3C![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3TCCLA 3TCCLA |
miR-7 | 55.0 | 60.0 |
| P6 | GAAGACLGTGLGA![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3T 3T![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3TC 3TC![[M with combining low line]](https://www.rsc.org/images/entities/char_004d_0332.gif) 3GGALAGA 3GGALAGA |
circRNA | 57.0 | 59.0 |
CuAAC click conjugation of ON1, ON2 with azides 3–5 was performed either under microwave conditions for 15 min at 60 °C (azide 3), or at ambient temperature for 24 h (azides 4 and 5), giving the desired products P1–P6 in high purity and yields of 74–83% (ESI†). Microwave conditions of the click reactions were applied for incorporation of monomer M1 in order to improve solubility of the azide 3. The products were characterized by ion-exchange (IE) HPLC and MALDI MS (ESI, Table S1†). Incorporation of the fluorescent monomers into ON1, ON2 was further confirmed by characteristic bands in the visible region of the corresponding absorbance spectra (Table 2; ESI, Table S4 and Fig. S1†). Notably, hybridization had an influence on the shape and intensity of absorbance curves which implied interaction of the dyes with microenvironment, e.g. dye aggregation and stacking interactions between the dyes and nucleobases.4,12,13
| # | λ absmax, (nm) | λ ex, (nm) | λ flmax (nm) | D | Φ f | FB | LOD, nM | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 19 °C | 37 °C | 19 °C | 37 °C | 19 °C | 37 °C | |||||
| a λ absmax, λex, λflmax, Φf, FB and LOD are maxima of absorbance, excitation wavelength, fluorescence maxima, fluorescence quantum yield, FB (fluorescence brightness) = Φf × εmax/1000, and limit of target detection values. Φf values are measured by a relative method (ESI) using perylene and oxazine 170 perchlorate as fluorescence standards. D is determined at 19 °C as a ratio of fluorescence intensities at fluorescence maximum of double-stranded complex to the corresponding single-stranded probe (452 nm for P1, P2, 530 nm (P3, P4), and 605 nm (P5, P6)). LODs were determined by series of target titration experiments described in ESI. | ||||||||||
| P1 | 423, 451 | 423 | 450 | 10.4 | 0.58 | 0.45 | 27 | 23 | 10 | 10 |
| P2 | 423, 451 | 423 | 488 | 5.6 | 0.47 | 0.40 | 34 | 32 | 10 | 10 |
| P3 | 480, 506 | 480 | 526 | 2.1 | 0.90 | 0.96 | 102 | 104 | <5 | <5 |
| P4 | 482, 506 | 480 | 528 | 6.9 | 0.90 | 0.99 | 133 | 138 | <5 | <5 |
| P5 | 549, 584 | 580 | 605 | 1.0 | 0.23 | 0.23 | 32 | 32 | 10 | 10 |
| P6 | 546, 585 | 580 | 607 | 1.1 | 0.18 | 0.18 | 30 | 30 | 10 | 10 |
Hybridization of the prepared probes with target DNA and RNA strands was studied using Tm and circular dichroism (CD) experiments in a medium salt phosphate buffer (Table 1, ESI, Fig. S2, S3 and Table S3†). For all the modified probes S-shape of the melting curves and the characteristic CD signal of an A/B type duplex confirmed successful adoption of the dyes within the duplexes (ESI, Fig. S2 and S3†).11 Additional LNA nucleotides improved binding affinities of the modified probes in spite of 5–9 °C destabilizing effect of monomer UP (Table 1, Tm for ON1–ON4 compared to unmodified references). Perylene-labelled probes P1, P2 showed high binding affinity towards complementary DNA/RNA (Tm 64–68 °C), which is most likely caused by additional stacking interactions provided by the polyaromatic hydrocarbons within the double stranded complexes.10 Furthermore, rhodamine-modified probes P3–P6 showed superior binding affinity towards RNA in comparison to DNA targets, indicating at better adoption of the bulky rhodamine fluorophores within the corresponding DNA–RNA hybrids (Tm 55–57 °C vs. 59–62 °C for the duplexes with DNA and RNA targets, respectively). However, incorporation of the fluorophores within monomers M2, M3 had a negative effect on Tm values compared to UP-modified and LNA/DNA references (Tm decrease ∼ 5 °C). All the probes discriminated a single mismatch in the central positions of DNA and RNA targets by −3.5 to 8 °C and −1 to 8 °C, respectively (ESI, Fig. S3†). Notably, the Tm values were comparable to those for 5′- and internally labeled LNA/DNA fluorescent probes obtained from commercial suppliers (ESI, Tables S2 and S3†). Finally, since all the duplexes having single-nucleotide mismatches were formed at the temperature above 19 °C, all of them were subjected to the fluorescence studies described below.
Fluorescence detection of hybridization with complementary miRNA, circRNA and their cDNA analogues was studied by in vitro experiments at ambient (19 °C) and elevated temperature (37 °C). The latter was done in order to evaluate potential of P1–P6 for in vivo applications. The in vitro assay using P1–P6 was characterized by series of photophysical and diagnostic parameters: discrimination values (D), fluorescence quantum yields (Φf), fluorescence brightness (FB) and limit of target detection (LOD) values (Table 2; ESI, Table S4 and Fig. S4†)10 In order to efficiently detect a target in vivo, D value of ≥10 is preferred. Perylene-labelled probes P1, P2 displayed high D values (up to 10.6), accompanied by high fluorescence quantum yields and high FBs at both 19 °C and 37 °C. The probes P3, P4 showed the highest Φf and FB values (up to 0.99 and 138, respectively), also at elevated temperature. High FBs resulted in low LOD values for P1–P4 (<5–10 nM), which is beneficial for diagnostic applications of the probes.5 However, low D values make them not applicable for diagnostic settings. Finally, P5, P6 showed similar FB and LOD values compared to those observed for P1, P2, and lower D values between single-stranded conjugates and corresponding complexes with miR-7 and circRNA (Table 2). The observed quenching of fluorescence within single-stranded P1–P4 most likely results from aggregation of the hydrophobic dyes surrounded by aqueous media, since both extinction coefficient and ratio of visible absorbance bands I/II of P1–P4 vary for the single strands and duplexes (ESI, Fig. S1†).13 In turn, as suggested by altered ratio of absorbance bands and increased extinction, hybridization results in positioning of the dyes in a less hydrophilic environment, resulting in the fluorescent signal.
Next, none of the commercially available 5′- or internally labeled LNA/DNA probes R1–R8 used in this study showed sensitivity of fluorescence to single-nucleotide mismatch in the RNA/cDNA targets (D 0.6–1.5; ESI, Fig. S5;†R1–R8 contain fluorophores with similar optical properties to M1–M3). On the contrary, “clickable” probes P1–P6 showed moderate fluorescence mismatch discrimination in cDNA targets (i.e. only numerous positions were discriminated; data not shown), and efficient discrimiation in most of the examined miR-7 and circRNA targets by quenching of fluorescence (e.g. D 0.1–0.7 for MT1–MT8 and probes P1, P3, P5; ESI, Tables S4, S5 and Fig. S5†). Overall, four duplexes out of forty eight studied showed low mismatch discrimination (D 0.9–1.6 by P4: MT1/MT3/MT8 and P2: MT7; ESI, Table S4†). This implies that the performed internal incorporation of monomers M1–M3 with expanded aromatic π-electron systems results in the probes which are in general potent to sense minor changes in microenvironment such as single-nucleotide mismatches.
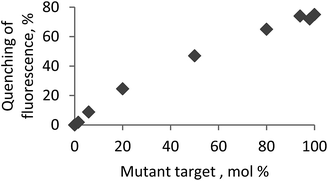
A critical challenge for detecting single-nucleotide polymorphisms (SNPs) in natural RNA targets is that most often a mutant genotype is present at a very low concentration with respect to the wild-type variant.14 Therefore, as the final aspect, biologically relevant mixtures of a wild-type and mismatched circRNA targets were analysed using the selected bright probe P4 and circRNA target MT13 containing A → G mismatch in the central position (Fig. 2; ESI, Tables S3 and S7†). Fluorescence of fully matched duplex P4–MT13 consistently decreased upon increasing concentration of the mismatched complex which allows accurate estimation of the mismatch abundance in the corresponding analyte. Finally, high base specificity of fluorescence signal is an additional advantage of the prepared probes in genotyping of natural RNA.14
Conclusions
In this work the rationally designed “clickable” probes are demonstrated to be promising tools in RNA detection by fluorescence. The background fluorescence of single strands was significantly reduced, while upon formation of complexes with target miR-7 and circRNA emission increased giving remarkably high quantum yields (Φf up to 0.99) and, therefore, low limit of detection values (<5 nM in solution). Importantly, this was achieved by internal modification of the LNA/DNA oligonucleotides providing additional thermal stability and selectivity of the probes towards the target. Accompanied by simple design and robust CuAAC synthetic route, simple purification and high yields of the products, effective target binding, low single-strand background signal (D up to 10.6) and consistent specific fluorescence, the “clickable” probes could become a reliable platform for detection of natural RNA.Acknowledgements
The authors acknowledge financial support from The Sapere Aude programme of The Danish Council for Independent Research. Lumiprobe LLC is acknowledged for providing azides 3–5.Notes and references
- P. J. Santangelo, E. Alonas, J. Jung, A. W. Lifland and C. Zurla, Methods Enzymol., 2012, 505, 383 CAS.
- A. H. El-Sagheer and T. Brown, Acc. Chem. Res., 2012, 45, 1258 CrossRef CAS.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879 CrossRef CAS PubMed.
- M. M. Rubner, C. Holzhauser, P. R. Bohländer and H.-A. Wagenknecht, Chem.–Eur. J., 2012, 18, 1299 CrossRef CAS PubMed; I. K. Astakhova and J. Wengel, Chem.–Eur. J., 2013, 19, 1112 CrossRef PubMed; I. K. Astakhova and J. Wengel, Acc. Chem. Res., 2014, 47, 1768–7177 CrossRef PubMed.
- D. D. Nolting, M. L. Nickels, N. Guo and W. Pham, Am. J. Nucl. Med. Mol. Imaging, 2012, 2, 273 Search PubMed , and references therein.
- N. Bushati and S. M. Cohen, Annu. Rev. Cell Dev. Biol., 2007, 23, 175 CrossRef CAS PubMed.
- T. B. Hansen, et al. , Nature, 2013, 495, 384 CrossRef CAS PubMed; G. Blandino, et al. , FEBS Lett., 2014, 588, 2639–2652 CrossRef PubMed.
- S. Memczak, et al. , Nature, 2013, 495, 333 CrossRef CAS PubMed.
- M. W. Hentze and T. Preiss, EMBO J., 2013, 32, 923 CrossRef CAS PubMed.
- S. P. Sau and P. J. Hrdlicka, J. Org. Chem., 2012, 77, 5 CrossRef CAS PubMed; I. K. Astakhova, E. Samokhina, B. R. Babu and J. Wengel, ChemBioChem, 2012, 13, 1509 CrossRef PubMed.
- I. K. Astakhova, T. S. Kumar, M. A. Campbell, A. V. Ustinov, V. A. Korshun and J. Wengel, Chem. Commun., 2013, 49, 511 RSC; A. S. Jørgensen, P. Gupta, J. Wengel and I. K. Astakhova, Chem. Commun., 2013, 49, 10751 RSC.
- For example, A. Chowdhury, S. Wachsmann-Hogiu, P. R. Bangal, I. Raheem and L. A. Peteanu, J. Phys. Chem. B, 2001, 105, 12196 CrossRef CAS.
- F. Würthner, T. E. Kaiser and C. R. Saha-Möller, Angew. Chem., Int. Ed., 2011, 50, 3376 CrossRef PubMed.
- Y. M. D. Lo, et al. , Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13116 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of synthesis, purification and characterization of the oligonucleotides, quantum yields and limit of target detection calculations, and spectral and photophysical data on targeting complementary and singly-mismatched DNA/RNA targets. See DOI: 10.1039/c4ra07165d |
| This journal is © The Royal Society of Chemistry 2014 |

![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) A UCC ACG UCU UC); mismatched nucleotide (replacement A → G) is underlined.
A UCC ACG UCU UC); mismatched nucleotide (replacement A → G) is underlined.