Feasibility study on anaerobic biodegradation of azo dye reactive orange 16
Mohammad Zain Khanab,
Satyendra Singha,
T. R. Sreekrishnana and
S. Z. Ahammad*a
aDepartment of Biochemical Engineering & Biotechnology, Indian Institute of Technology Delhi, Hauz Khas, New Delhi 110016, India. E-mail: zia@dbeb.iitd.ac.in
bDepartment of Chemistry, Aligarh Muslim University, Aligarh, Uttar Pradesh, India
First published on 12th September 2014
Abstract
Anaerobic digestion of textile azo-dyes is very effective and widely used since it is cost-effective and energy efficient. The present study deals with the anaerobic degradation of reactive orange 16 (RO 16, an azo-dye) using mixed microbial culture. 80 mL each of three different concentrations of RO 16 (100, 200 and 300 ppm) were taken in 150 mL serum vials containing 20 mL of mixed microbial culture and studied periodically. HPLC and UV data revealed that more than 90% of the color was removed within the very first week of the reactor startup. A high COD removal efficiency (≥80%) was achieved after the steady state. Methane and VFAs were produced, and monitored by Gas chromatography. The pH of the medium was slightly acidic favoring methanogenic activity. The diversity of the microbial community was studied by denaturing gradient gel electrophoresis (DGGE) of the polymerase chain reaction (PCR) amplified products of the bacterial and archeal 16S rRNA and the results showed the presence of significant population of acetogens as well as methanogens in the reactor. Quantitative real time PCR (qPCR) was used for the quantitative analysis of some major genera. This study showed that strategic operation of the anaerobic digester can be a viable option for effective decolorization of a complex substrate resulting in energy (biogas) generation.
1 Introduction
The textile industry accounts for two-thirds of the total dyestuff market and azo-dyes constitute the largest class of synthetic dyes in commercial applications.1 During the dyeing process, approximately 10–15% of the dyes used are released into wastewater.2,3 They are complex aromatic xenobiotic compounds, very recalcitrant towards biological degradation and are responsible for intense color, high water solubility, toxicity and resistant to degradation under natural conditions.4,5 The high color content of these dyes inhibits photosynthesis in aquatic plants and algae through absorption of light.6 Although textile effluents contain a variety of dyes with high organic content, its composition varies significantly depending upon the geographical region, type of industry and climatic conditions.1Although, several methods such as filtration, coagulation, adsorption (activated carbon) and chemical flocculation are used and been tried to decontaminate textile effluents containing azo dyes in order to achieve decolorization,7 but they expensive and sometimes causes secondary pollution.8 On the other hand, biological treatment methods are attractive due to their cost effectiveness, diverse metabolic pathways and versatility of microorganisms.9,10
Moreover, the treatment of dissolve chemical oxygen demands through biological processes (activated sludge process) requires energy intensive traditional technologies.11,12 These conventional processes usually consume 1 kW h of electricity and produce 0.4 kg of sludge per kg of oxidized COD which required further treatment its before disposal.13 Hence, anaerobic biological treatment processes are an excellent alternative in the sense that they can generate energy in the form of biogas (methane, carbon dioxide etc.) which can off-set the treatment cost.14 Anaerobic treatment involves four different stages viz. hydrolytic phase, acidogenic phase, acetogenic phase and methanogenic phase.15 Generally, the desired goal in anaerobic system is to achieve the same rate of formation of acids (acidogenic and acetogenic phase) as that of the rate of consumption of these acids for formation of methane.16 However, in most cases, the rate of acid formation is faster leading to the accumulation of acids and a drop in pH, causing inhibition of methanogenic activity.17,18 Presence of sulphate reducing bacteria may also inhibit the methanogenic activity.
During the methanogenic phase, carbon dioxide and/or acetic acid acts as terminal electron acceptors for methane production. However, the microorganisms involve in this stage are very substrate specific as some of them utilizes acetate or formate while some others prefer CO2 or CO as substrate. The optimum pH for growth of these microorganisms is 6 to 7.19
Following are the common reaction in methanogenic phase19,20
| CH3COOH → CH4 + CO2 (acetoclastic methanogens) |
| 4H2 + CO2 → CH4 + 2H2O (H2 utilizer methanogens) |
In case of anaerobic degradation of azo-dyes, azo-reductase plays a major role by cleaving the azo bond and yielding aromatic amines.21,22 The aromatic amines are generally recalcitrant towards anaerobic conditions with an exception of few aromatic amines containing hydroxyl and/or carboxyl group.23,24 However, long retention times could cause mineralization of metabolites of some azo-dyes even under anaerobic conditions.25 Razo-Flores and co-workers showed a complete anaerobic mineralization of the azo-dye azodisalicylate under methanogenic conditions and claimed that it was firstly reduced to the corresponding aromatic amine 5-aminosalicylic acid which later mineralized completely under anaerobic conditions.26
Ar–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–Ar′ + 4e− + H+ → Ar–NH2 + Ar′–NH2 N–Ar′ + 4e− + H+ → Ar–NH2 + Ar′–NH2 |
The main aim of the present study was to check the feasibility of degrading reactive orange 16 dye (RO 16) as sole carbon source (without any co-substrate) under strictly anaerobic conditions and to characterize the microbial community using molecular biology techniques. Performance of the anaerobic reactor was estimated by monitoring VFAs (volatile fatty acids) and biogas (methane) produced during RO 16 degradation. Along with the chemical parameters, microbial community responsible for effective degradation was also estimated. Culture independent, DNA based, advanced molecular microbiology tools such as DGGE and qPCR were used to determine the microbial community profile and identify the key microorganisms responsible for effective degradation of RO 16. Data obtained from the study could be used to develop strategy for operating the anaerobic digester to degrade azo dyes at optimum reactor performance.
2 Experimental
2.1 Chemicals
All the chemicals used were of analytical grade and procured from Merck India. The azo-dye (reactive orange 16) was obtained from the Department of Textile Technology, IIT Delhi and was manufactured by Vipul Dyes, India.2.2 Inoculum
The anaerobic seed sludge for inoculation was taken from an anaerobic digester at Okhla Sewage Treatment Plant, New Delhi, India. The concentration of volatile suspended solids in the initial sludge was 3 g L−1. The anaerobic sludge was characterized by estimating the specific methane production rate. A synthetic media was used to estimate the methanogenic activity of the anaerobic sludge in batch reactors. Composition of the salt media used in all sludge activity tests was –NH4Cl 0.85 g L−1, KH2PO4 0.136 g L−1, K2HPO4 0.234 g L−1, MgCl2·6H2O 0.084 g L−1, FeCl3 0.05 g L−1 and Yeast extract 0.34 g L−1.27 Along with the salt solution, 20 mM acetate was used in batch reactors having 0.2, 0.4, 0.8 and 1 gm of the anaerobic sludge to test the activity of acetoclastic methanogens. Also, 20 mM formate in the salt solution was used in batch reactors having 0.2, 0.4, 0.8 and 1 gm of the anaerobic sludge to test the activity of hydrogenotrophic methanogens present in the sludge. The solution pH was adjusted to 6.8–7.0 using dilute HCl. The vials were sparged with nitrogen gas and kept on shaker incubator at 37 °C, 100 rpm for 8 weeks and methane production was monitored periodically. The specific methanogenic activity (mM methane/gVSS/d) was calculated in terms of mM methane produced per gram of volatile solids per day. All the experiments were carried out in triplicate. The methane production rate of the sludge was 1.8 mM CH4/gVSS/d. Control vials without any substrate were also used to consider the background methane production data.2.3 Experimental set up for dye degradation
80 mL each of three different concentrations (100, 200 and 300 ppm) of RO 16 azo-dye were taken in 150 mL serum vials containing 20 mL of anaerobic sludge. The vials were sparged with nitrogen to remove any oxygen and to maintain strict anaerobic conditions before starting the experiment. The experiment was conducted in triplicate for each condition. Controls for each concentration without anaerobic sludge and biomass-controls containing only the anaerobic sludge without any carbon source were also maintained in triplicate during the experiment. Biomass-controls were used to correct background methane production by the sludge. All the vials were kept on a shaker incubator at 37 °C and 100 rpm for 8 weeks. Samples were collected and analyzed after every week.2.4 Analytical methods
COD was estimated using closed reflux method and spectrophotometer in accordance with the Standard methods.28 Methane and carbon dioxide formed during methanogenic phase were estimated using a Gas Chromatograph (AIMIL NUCON 5700 Series, India) with a 6-feet long Porapack-Q column and a thermal conductivity detector (TCD). Hydrogen was used as carrier gas. Standardization of the instrument was done using standard calibration mixture containing 50% methane and 50% carbon dioxide (Sigma gases, India). The conditions maintained for methane detection were: Injector temperature 80 °C; Oven temperature 50 °C; Detector temperature 80 °C; TCD Current 80 mA. The concentration of dye and decolorization rate was determined using HPLC (SHIMADZU LC-20 AD, Japan) having Eclipse XDB-C18 column (4.6 × 150 mm) (Agilent, USA) with 100% pure methanol used as mobile phase. The flow rate for the mobile phase was maintained at 1.0 mL min−1. The dye was detected by UV-visible detector (SHIMADZU LC-20 AD, Japan) at 422 nm wavelength and quantified on the basis of area and retention time of the respective standards. The results of HPLC were also supported by UV-visible spectra of the treated samples taken in the wavelength range 190–700 nm using UV-visible spectrophotometer (SPD-20A, Japan).The volatile fatty acids (acetate, propionate and butyrate's) formed during the process were monitored by Gas-Liquid Chromatograph (GLC) (AIMIL NUCON 5765 series, India) equipped with Flame Ionization Detector (FID) and a 6-feet long Chromosorb 101 column. The following conditions were maintained for estimating the VFAs: detector temperature 200 °C; injector temperature 195 °C; oven temperature 180 °C. Nitrogen was used as the carrier gas and a mixture of hydrogen and air were used to sustain the flame in the detector. Calibration of the instrument was done using standard solutions of pure VFAs in distilled water.
2.5 Scanning electron microscopy
The surface morphology and the diversity of the anaerobic microbial community during RO16 degradation were studied using a scanning electron microscope (ZEISS EVO Series 50, USA). The anaerobic culture was prepared for SEM analysis by fixing it with 20% glutareldehyde for an hour followed by successive washing with ethanol (5, 10, 20, 40, 60, 80 and 100%). Dried samples were taken for SEM analysis and were coated with gold before microscopic imaging.2.6 DNA extraction and amplification by PCR
The microbial diversity of the mixed microbial culture used in the present study was characterized by various molecular biological techniques such as polymerase chain reaction (PCR), denaturing gradient gel electrophoresis (DGGE) and quantitative real time polymerase chain reaction (qPCR).DNA was isolated using the FAST DNA SPIN kit from MP Biomedicals, USA. The DNA was isolated in accordance with the method given by Ahammad et al.29 The concentration of DNA in the isolated samples was around 40 ng μL−1. In order to get a more clear DGGE analysis for archaea, a nested PCR strategy was employed in which the first round PCR (Bio-Rad, C1000 Thermocycler, USA) was performed with primer set PRA46f and PRA1100r of amplicon size of 1054 bp. The following thermal cycling was used: Initial denaturation at 92 °C for 3 min followed by 30 cycles of 92 °C for 1 min, annealing at 55 °C for 1 min, with a final elongation step at 72 °C for 7 min. In the second round, the PCR products of first round were re-amplified with a set of universal primers PARCH340f-GC and PARCH519r with the following thermal cycling programme: Initial denaturation at 95 °C for 3 min followed by 30 cycles of 92 °C for 1 min, annealing at 55 °C for 1 min, with a final elongation step at 72 °C for 7 min.
In the first round of amplification the following recipe was used for making 25 μL PCR reaction mix. PCR master mix (Bioline, UK) 12.5 μL, H2O 9.5 μL, forward primer PRA46f 0.5 μL, reverse primer PRA1100r 0.5 μL, template DNA 2 μL. In the nested PCR, 50 μL reaction mix was prepared using 25 μL PCR master mix (Bioline, UK), 21 μL H2O, 1.0 μL forward Primer (P340f), reverse Primer (P519r-GC) having added GC clamps 1.0 μL, template DNA 1 μL (amplified DNA from the first round of PCR).
Primers UNIBACT341f-GC and UNIBACT518r were used for amplification of bacterial (V3) 16S rRNA gene fragment. The following thermo cycler programme was used for amplification of bacterial V3 region: 95 °C for 3 min initial denaturation followed by 35 cycles of 95 °C for 1 min, a touchdown protocol was used at annealing step with 65 to 55 °C for 30 second with a decrement of (−0.5 °C/step), elongation steps 72 °C for 1 min and 72 °C for 7 min for final elongation. The sizes of amplicon was checked by electrophoresis in 1.5% (wt/V) agarose gel stained with ethidium bromide. The list of primers used for PCR is provided in Table 1.
| Name | Target group | Function | Sequences (5′–3′) | References |
|---|---|---|---|---|
| PRA46f | Archaea | Forward | (C/T)TAAGCCATGC(G/A)AGT | 34 |
| PRA1100r | Reverse | (T/C)GGGTCTCGCTCGTT(G/A)CC | 34 | |
| PARCH340fGC | Archaea, V3 region | Forward | CCCTACGGGG(C/T)GCA(G/C)CAG CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG | 34 |
| PARCH519r | Reverse | TTACCGCGGC(G/T)GCTG | 34 | |
| UNIBACT341fGC | Bacteria, V3 region | Forward | ACTCCTACGGGAGGCAGCAG CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG | 35 |
| UNIBACT518r | Reverse | ATTACCGCGGCTGCTGG | 30 |
2.7 Microbial characterization of the sludge using DGGE
Denaturing gradient gel electrophoresis (DGGE) technique was used to estimate the community profile of the sludge samples and to distinguish between the initial (startup) and final (after 8 weeks of operation) sludge obtained in the reactor after anaerobic digestion. DGGE was performed in accordance with the protocol given by Muyzer et al.30 Bio-Rad D Code Universal Gene Mutation System (Bio-Rad Laboratories, Hercules, CA, USA) was used for running the gel. The PCR amplified products of the second round of the nested PCR were loaded on 8% polyacrylamide gels in 1% TAE (20 mmol L−1 Tris, 10 mM acetate and 0.5 mM EDTA pH 7.4) and a gradient of 45–60% was maintained. The gel was run at 60°C and 70 V for 16 h. Immediately after the gel electrophoresis; the plates were removed from the D-Code assembly and soaked in SYBR gold for 30 min for staining the gels. The stained gels were photographed in a Gel Documentation imaging system (Bio-Rad Laboratories, Hercules, CA, USA). Band pattern obtained were subjected to digital analysis. The intensities of the bands were analysed by Gel Doc XR+, Image LabTM 2.0 (Bio-Rad, Hercules, CA, USA).2.8 Estimation of microbial abundance using quantitative PCR (qPCR)
Prior to qPCR analysis, DNA extracts were diluted with molecular-grade water to minimize the influence of possible PCR inhibitors present in the extracted DNA. Different dilutions of DNA were checked and inhibition free PCR amplification was obtained with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted DNA samples. The diluted DNA samples were used for all qPCR analysis using a BioRad CFX C1000 (Hercules, CA, USA) qPCR system to target 16S rRNA, SRB and methanogens. Each 10 μL reaction mixture contained: 3 μL of template DNA, 0.5 μL of each primer (10 pmol μL−1), 1 μL nuclease free water and 5 μL qPCR reagent (SsoFast EvaGreen Supermix, Biorad, USA). Primers used in the qPCR reactions are provided in Table 2 and the qPCR programmes used were as follows: Eubacteria: initial enzyme activation at 95 °C for 5 min, denaturation at 95 °C for 45 s, annealing at 60.5 °C for 45 s and extension step at 72 °C for 45 s. The reaction was continued for another 39 cycles. Methanomicrobiales (MMB); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 62 °C for 5 s. The reaction was continued for another 45 cycles. Methanosarcinaceae (MSC) and Methanosaetaceae (MST); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 60 °C for 5 s. The reaction was continued for another 45 cycles. Methanobacteriales (MBT) and Methanococcales (MCC); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 60 °C for 5 s. The reaction was continued for another 45 cycles. Finally, SRB; initial enzyme activation at 94 °C for 5 min, denaturation at 94 °C for 45 s, annealing and extension at 59 °C for 60 s and extension step at 72 °C for 45 s. The reaction was continued for another 39 cycles.
10 diluted DNA samples. The diluted DNA samples were used for all qPCR analysis using a BioRad CFX C1000 (Hercules, CA, USA) qPCR system to target 16S rRNA, SRB and methanogens. Each 10 μL reaction mixture contained: 3 μL of template DNA, 0.5 μL of each primer (10 pmol μL−1), 1 μL nuclease free water and 5 μL qPCR reagent (SsoFast EvaGreen Supermix, Biorad, USA). Primers used in the qPCR reactions are provided in Table 2 and the qPCR programmes used were as follows: Eubacteria: initial enzyme activation at 95 °C for 5 min, denaturation at 95 °C for 45 s, annealing at 60.5 °C for 45 s and extension step at 72 °C for 45 s. The reaction was continued for another 39 cycles. Methanomicrobiales (MMB); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 62 °C for 5 s. The reaction was continued for another 45 cycles. Methanosarcinaceae (MSC) and Methanosaetaceae (MST); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 60 °C for 5 s. The reaction was continued for another 45 cycles. Methanobacteriales (MBT) and Methanococcales (MCC); initial enzyme activation at 98 °C for 3 min, denaturation at 98 °C for 2 s, annealing and extension at 60 °C for 5 s. The reaction was continued for another 45 cycles. Finally, SRB; initial enzyme activation at 94 °C for 5 min, denaturation at 94 °C for 45 s, annealing and extension at 59 °C for 60 s and extension step at 72 °C for 45 s. The reaction was continued for another 39 cycles.
| Name | Target group | Function | Sequences (5′–3′) | References |
|---|---|---|---|---|
| Bact338 | Eubacteria | Forward | ACTCC TACGG GAGGC AG | 36 and 37 |
| BAct1046 | Reverse | CGACARCCATGCANCACCT | ||
| MSC380F | Methanosarcinaceae | Forward | GAA ACC GYG ATA AGG GGA | 36 |
| MSC828R | Reverse | TAG CGA RCA TCG TTT ACG | ||
| MST702F | Methanosaetaceae | Forward | TAA TCCT YGA RGG ACC ACC A | 36 |
| MST862R | Reverse | CCT ACG GCA CCR ACM AC | ||
| MMB282F | Methanomicrobiales | Forward | ATC GRT ACG GGT TGT GGG | 36 |
| MMB832R | Reverse | CAC CTA ACG CRC ATH GTT TAC | ||
| MBT857F | Methanobacteriales | Forward | CGWAG GGAAG CTGTT AAGT | 36 |
| MBT1196R | Reverse | TACCG TCGTC CACTC CTT | ||
| MCC495F | Methanococcales | Forward | TAAGG GCTGG GCAAG T | 36 |
| MCC832R | Reverse | CACCT AGTYC GCARA GTTTA | ||
| DsrA-F | Sulfate reducing bacteria (dsrAB gene) | Forward | ACS CAC TGG AAG CAC GGC GG | 38 |
| DsrA-R | Reverse | GTG GMR CCG TGC AKR TTG G |
3 Results and discussion
3.1 COD removal in the batch reactor
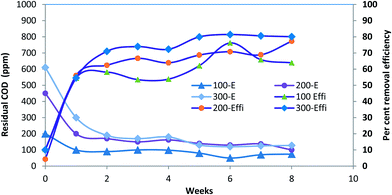
The residual COD in the batch reactors containing reactive orange 16 in synthetic media as well as the percent COD removal are shown in Fig. 1. The COD was contributed by only azo-dye as no growth substrate was used in the present work. The COD reduction in the dye-control samples (containing only dye substrate but no sludge) is minimal and owing to vaporization losses only and no degradation was observed. In case of the sludge-control samples (containing anaerobic sludge only, without any dye substrate) the COD values were nearly zero, since the conditions were strictly anaerobic. The COD for all the test samples shows a gradual decreasing trend, which finally stabilizes below 100 ppm in case of batch reactors treating 100 and 200 ppm of RO 16 while the final COD values in case of reactor treating 300 ppm of RO 16 were around 180 ppm. The higher residual COD concentrations in case of the reactor treating 300 ppm of RO 16 are due to microbial inhibition caused by high substrate concentrations itself.19 As far as the COD removal efficiency is concerned, it was low initially, but improved substantially after few weeks of operation in the batch reactors. All the test reactors achieved removal efficiency of 80% and above except the third reactor (reactor degrading 300 ppm of RO 16) due to substrate inhibition effects as mentioned above. The COD removed was stoichiometrically converted to VFAs which are finally transformed in to biogas by the action of methanogens.However, in some reports available in the literature, a low COD removal is observed owing to the formation of aromatic amines as a reduction product of azo-dyes which are recalcitrant towards biodegradation under anaerobic conditions.31,32
3.2 Dye degradation in the reactor
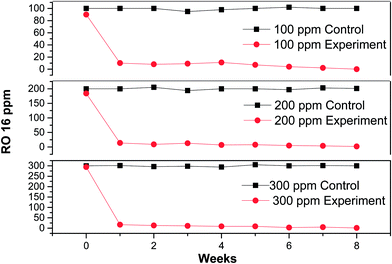
The dye solution shows four peaks (two in the UV region at 250, 290 nm and two in the visible region at 386 and 495 nm) attributing to the different chromophoric groups. The absorbance peaks at 250 and 290 nm are due to the benzene and naphthalene rings, and the absorbance peaks at 386 and 490 nm are due to the azo linkage of RO 16 and responsible for color.23,33 The merged absorption spectra of different samples are shown in Fig. 2. The lowering of absorbance peaks indicates degradation and subsequent removal of RO 16 as a result of anaerobic degradation.In order to verify UV-Visible absorbance data, residual RO 16 concentrations were further monitored with the help of HPLC system. The data revealed complete decolorization of the dye within the first week of inoculation (Fig. 3). From the second week onwards, the plots for all the three experimental reactors were identical to the control plot showing no color and claiming that a high decolorization rate was achieved during the study. Moreover, a number of reports available in literature show the possibility of formation and accumulation of aromatic amines as degradation products of azo-dyes e.g. Mendez-Paz et al.31 yields sulphanilic acid and 1-amino-2-naphthol during anaerobic degradation of Acid Orange 7 azo-dye in which 1-amino-2-napthol was not detected likely because of its low stability. Ahmad et al.32 reported the formation of two aromatic amines p-amino diphenylamine and 4-aminobenzenesulfonic acid from the reduction of Acid Yellow-36, which are toxic and mutagenic in nature. In case of such recalcitrant and toxic aromatic amines, a combined anaerobic-aerobic process is preferred for complete removal.
However, in the present case, no peak corresponding to aromatic amines was observed in the UV spectra possibly due to the long retention time (almost 60 days) in the reactors resulted in degradation and subsequent removal of the aromatic amines formed during reductive cleavage of azo-dyes.25,26 The formation of VFAs (Fig. 4) and biogas also indicate mineralization of aromatic amines since no co-substrate was used in the present study.
3.3 Volatile fatty acids formation
VFAs were formed during the acetogenic phase of the anaerobic digestion and comprises mainly of acetic acid, propionic acid and butyric acid. Long chain acids are being oxidized by acetogenic bacteria resulting in VFAs.16 Hydrogen and carbon dioxide are also formed in this phase and there is a balance between acetogenic bacteria producing hydrogen and methanogens consuming hydrogen. It is also known that acetate formation depends upon the syntrophic association between acetogens and methanogens.15,16 If acetogens are more active, more acetate will accumulate in the reactor leading to a fall in the pH and vice versa.VFA formed during the anaerobic digestion in the reactors showed presence of acetate and propionate however, no butyrate was observed in any of the reactors (Fig. 4). The acetate and propionate act as electron donor and may support the activity of sulphate reducing bacteria (converting SO42− to S2−). At the end of the experiment, no residual acetate as well as propionate were found since all the acids were converted to methane and carbon dioxide by the acetoclastic methanogenic community.
3.4 Production of biogas
Methane and carbon dioxide were produced during RO 16 degradation under anaerobic conditions. The volumetric production of the biogas was transformed to millimoles per liter (mill molar ‘mM’) and presented in Fig. 5. The production was low initially as the system was dominated by acidogenic microbial communities. However, at later stages the gas production improved substantially and finally at the end of the operation the rate again slowed down due to the substrate limitations.25 It was observed that a sufficient feast to famine ratio (microorganism to substrate) must be maintained for large amount of methane production and whenever this ratio is low, methane production is low. | ||
| Fig. 5 Methane and CO2 profiles obtained during different weeks of the experimental stage in the batch reactors. | ||
Fig. 5 clearly shows that reactor containing 200 ppm RO 16 produced highest amount of methane and CO2 due to high substrate availability (and high COD removal). However, the high concentration of RO 16 in case of 300 ppm causes inhibition of microbial growth leading to lower methane production.19 In case of control samples containing anaerobic culture with no substrate, an endogenous metabolism occurred wherein the microbial communities start consuming the dead microorganisms resulting in methane production. Hence, it can be concluded that methane production can be obtained during degradation of toxic substrate like azo-dye and the energy can be utilized for heating the digester or for any other useful purpose.
3.5 Surface characterization by SEM analysis
The surface structures of the anaerobic sludge shows rod shaped bacteria are predominant with some cocci and diatoms (Fig. 6). The change in color of the final sludge and initial inoculum is due to the feeding of the RO 16 which leads to the accumulation of a few new strains, along with some amine derivatives. The sludge morphology is quite different in both the cases. SEM images provide information only on the surface morphology and presence of diverse class of microbial species. However, the actual identification, characterization and quantification of the microbial species were done with the help of DGGE and qPCR techniques. | ||
| Fig. 6 SEM images of the anaerobic culture [1] Initial inocula and [2] Final sludge (after 8 weeks of operation). | ||
3.6 Microbial community analysis using PCR-DGGE
The 16S rDNA fragments of the bacteria as well as archea present in the mixed microbial consortia in the reactors were amplified by a nested PCR strategy and the PCR products were further analyzed using DGGE technique. Eight weeks long azo-dye treating archeal enriched anaerobic microbial culture was analyzed for the bacteria as well as archea. The gel pictures having different bands obtained during DGGE analysis are shown in Fig. 7A & B. Different bands on the DGGE gel represented different bacterial genotypes. A total of 12 different bands were observed in the bacterial community present in the initial inoculum and in the samples collected at different time periods during the experiment. It was observed that the intensity of almost all bands increased in the samples collected at different time intervals compared to the initial inoculum. Increase in band intensity indicates the enrichment of bacterial community in the reactor during the experiment. It also demonstrates that the azo-dye degradation has favored the growth of the bacterial community (Fig. 7A).However, in case of the archaeal community, the inoculum showed 11 distinct but faint bands out of 17 bands in azo dye treating consortia (Fig. 7B). The band numbers 15, 16 & 17 were not properly resolved in inoculum as shown in Fig. 7B Lane 1 due to lower abundance of the belonging community, which further, got resolved and clearly appeared on consecutive weeks of incubation. Some new bands, 10th, 12th & 13th started getting enriched during the course of the experiment and they were not appeared in the inoculum. Since the DGGE is only a qualitative technique, it does not provide any evidence regarding the absolute abundance of the population of different microbial communities. It only provides information about the relative abundance of microbial communities and presence or absence of bands in different samples. But the analysis showed a clear indication of enrichment of certain communities in the bacterial and archaeal population. To estimate the specific community responsible for methane production, quantification of different microbial species was carried out using qPCR technique.
3.7 Analysis of abundance of key microorganisms in the reactor using quantitative PCR (qPCR)
The quantitative real time PCR technique provides information on the absolute abundance of gene of different species present in the microbial community in the samples collected during the course of the experiment (Fig. 8). The following major classes of methanogens were targeted with qPCR-Methanosarcina (MSC), Methanoseata (MST), Methanococcus (MCC), Methanobacterium (MBT) and Methanomicrobium (MMB) along with sulphate reducing bacteria (SRB).It can be clearly seen from the Fig. 8 that the total bacterial count (gene copy number per mL) has decreased in week 6 and 8. Thus, a sufficient food to microorganism ratio is essential for proper growth and to obtain activity of microorganisms. Hydrogenotrophic methanogens showed increased population compared to acetoclastic methanogens. Methanomicrobium and Methanococcus showed substantial increase in population while methanobacterium population did not increased much. The increased population of hydrogen utilizing bacteria in the reactors shows that hydrogen was primarily utilized for methane production. However, there was no significant change in the population of sulphate reducing bacteria. This also indicates that the SRB could not outcompete with the hydrogenotrophic methanogens. This microbial analysis also suggest that operation of the reactor by maintaining optimum conditions for hydrogen utilizing methanogens could improve the performance of the reactor.
4 Conclusion
The anaerobic degradation of RO 16 using mixed microbial culture was achieved with high COD removal efficiency. The VFAs (acetate and propionate) were produced by acetogens and transformed into methane and carbon dioxide by methanogens. The methane production was in good agreement with the COD removal. Although, methane production was good for all the test concentrations, the highest amount was produced in case of 200 ppm with decrease in production rates as we go further to 300 ppm due to substrate inhibition. The surface structures of the anaerobic sludge shows rod shaped bacteria are predominant with some cocci and diatoms. A nested PCR strategy was used successfully to amplify the gene sequence followed by DGGE to characterize the microbial community. The reactor was dominated by bacterial as well as archaeal (methanogenic) communities. Thus, degradation of toxic and complex substrates can be successfully achieved under anaerobic conditions with simultaneous methane production. The microbial community analysis indicated that the hydrogenotrophic methanogens are the key community for degrading the azo dye and production of methane. They can be used as a possible indicators for estimating the health of the anaerobic digesters degrading azo dye.Although a complete treatment of azo-dye was not guaranteed always under anaerobic condition due to the possible formation of bioaccumulative degradation products but the use anaerobic pre-treatment would definitely reduce the energy usage and improve treatment process economy compared to aerobic system alone. The use of anaerobic pre-treatment would definitely reduce the energy usage and improve treatment process economy compared to aerobic system alone. Substantial reduction of COD (up to 80%) in anaerobic reactor suggests an anaerobic followed by aerobic treatment systems would be the best option for complete removal of azo dye containing wastewater. Anaerobic reactors require more stringent maintenance and skilled operator compared to aerobic reactor mostly because of delicate nature of methanogens. Knowledge of microbial community present in the anaerobic reactor would help in managing the reactor and preventing the failure of the reactor. Microbial community profile data obtained from the anaerobic reactor would help to select appropriate operating strategy to be maintained in the anaerobic reactor to favour the key/indicative microorganisms, hydrogentrophic methanogens for degradation of azo dyes. The experiment shows a feasibility of using anaerobic treatment to degrade azo dyes as a sole source of carbon and energy and also indicates a strong possibility to use the technology in larger scale to achieve low-energy treatment strategy for treating textile wastewater containing azo dyes.
Acknowledgements
The authors are thankful to Department of Science & Technology, Government of India for providing financial support to carry the research (Grant number SB/FT/CS-037/2012).References
- P. C. Vandevivere, R. Bianchi and W. Verstraete, J. Chem. Technol. Biotechnol., 1998, 72, 289–302 CrossRef CAS.
- F. Elisangela, Z. Andrea, D. G. Fabio, R. de Menezes Cristiano, D. L. Regina and C. P. Artur, Int. Biodeterior. Biodegrad., 2009, 63, 280–289 CrossRef CAS PubMed.
- J. T. Spadaro, I. Isabelle and V. Renganathan, Environ. Sci. Technol., 1994, 28, 1389–1393 CrossRef CAS PubMed.
- H. Zollinger, Color Chemistry: Syntheses, Properties and Applications of Organic Dyes and Pigments, VHC Publishers, New York, 2nd edn, 1991 Search PubMed.
- A. Stolz, Appl. Microbiol. Biotechnol., 2001, 56, 69–80 CrossRef CAS.
- I. M. Banat, P. Nigam, D. Singh and R. Marchant, Bioresour. Technol., 1996, 58, 217–227 CrossRef CAS.
- P. R. Gogate and A. B. Pandit, Adv. Environ. Res., 2004, 8, 553–597 CrossRef CAS.
- J. Maier, A. Kandelbauer, A. Erlacher, A. C. Paulo and G. M. Gubitz, Appl. Environ. Microbiol., 2004, 70, 837–844 CrossRef CAS.
- A. Pandey, P. Singh and L. Iyengar, Int. Biodeterior. Biodegrad., 2007, 59, 73–84 CrossRef CAS PubMed.
- C. Aloius Togo, C. C. Z. Mutambanengwe and C. G. Whiteley, Afr. J. Biotechnol., 2008, 7, 114–121 Search PubMed.
- M. Z. Khan, P. K. Mondal and S. Sabir, J. Hazard. Mater., 2011, 190, 222–228 CrossRef CAS PubMed.
- M. Z. Khan, P. K. Mondal, S. Sabir and V. Tare, Bioresour. Technol., 2011, 102, 7016–7021 CrossRef CAS PubMed.
- K. Rabaey, N. Boon, M. Hofte and W. Verstraete, Environ. Sci. Technol., 2005, 39, 3401–3408 CrossRef CAS.
- F. Raposo, M. A. De la Rubia, V. Fernández-Cegrí and R. Borja, Renewable Sustainable Energy Rev., 2011, 16, 861–877 CrossRef PubMed.
- H. W. Yu, Z. Samani and G. Smith, Waste Manag., 2002, 22, 1–5 CrossRef CAS.
- S. Ghosh, Water Sci. Technol., 1991, 23, 1179–1188 CAS.
- J. V. Sachs, U. Meyer, P. Rys and H. Feitkenhauer, Water Res., 2003, 37, 973–982 CrossRef.
- R. Solera, L. I. Romero and D. Sales, Chem. Biochem. Eng. Q., 2002, 16, 25–29 CAS.
- F. P. Van der Zee, G. Lettinga and J. A. Field, Chemosphere, 2001, 44, 1169–1176 CrossRef CAS.
- F. P. Van der Zee and S. Villaverde, Water Res., 2005, 39, 1425–1440 CrossRef CAS PubMed.
- D. Mendez-Paz, F. Omil and J. M. Lema, Enzyme Microb. Technol., 2005, 36, 264–272 CrossRef CAS PubMed.
- D. Georgiou, C. Metallinou, A. Aivasidis, E. Voudrias and K. Gimouhopoulos, Biochem. Eng. J., 2004, 19, 75–79 CrossRef CAS PubMed.
- E. Franciscon, M. J. Grossman, J. A. R. Paschoal, F. G. R. Reyes and L. R. Durrant, SpringerPlus, 2012, 37, 1–10 Search PubMed.
- S. Kalyuzhnyi, V. Sklyar, T. Mosolova, I. Kucherenko, J. A. Russkova and N. Degtyaryova, Water Sci. Technol., 2000, 42, 363–370 CAS.
- M. Isik and D. T. Sponza, Enzyme Microb. Technol., 2007, 40, 934–939 CrossRef CAS PubMed.
- E. Razo-Flores, M. Luijten, B. A. Donlon, G. Lettinga and J. A. Field, Environ. Sci. Technol., 1997, 31, 2098–2103 CrossRef CAS.
- R. E. Speece, Anaerobic biotechnology for industrial wastewaters, Archae Press, Nashville, TN, USA, 1996, pp. 1–320 Search PubMed.
- APHA, Standard Methods for the Examination of Water and Wastewater, American Public Health Association, Washington, D.C., USA, 20th edn, 2002 Search PubMed.
- S. Z. Ahammad, R. J. Davenport, L. F. Read, J. Gomes, T. R. Sreekrishnan and J. Dolfing, RSC Adv., 2013, 3, 774–781 RSC.
- G. Muyzer, E. C. Waal and A. G. Uitterlinden, Appl. Environ. Microbiol., 1993, 59, 695–700 CAS.
- D. Mendez-Paz, F. Omil and J. M. Lema, Enzyme Microb. Technol., 2005, 36, 264–272 CrossRef CAS PubMed.
- R. Ahmad, P. K. Mondal and S. Q. Usmani, Bioresour. Technol., 2010, 101, 3787–3790 CrossRef CAS PubMed.
- J. Mitrovic, M. Radovic, D. Bojic, T. Andelkovic, M. Purenovic and A. Bojic, J. Serb. Chem. Soc., 2012, 77, 465–481 CrossRef CAS.
- L. Ovreas, L. Forney, F. L. Daae and V. Torsvik, Appl. Microbiol. Biotechnol., 1997, 63, 3367–3373 CAS.
- D. J. Lane, 16S/23S rRNA sequencing, in Nucleic acid techniques in bacterial systematic, ed. E. Stackebrandt and M. Goodfellow, John Wiley & Sons Ltd., West Sussex, United Kingdom, 1991, pp. 115–175 Search PubMed.
- Y. Yu, C. Lee, J. Kim and S. Hwang, Biotechnol. Bioeng., 2005, 89, 670–679 CrossRef CAS PubMed.
- J. A. Huber, D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield and M. L. Sogin, Science, 2007, 318, 97–112 CrossRef CAS PubMed.
- R. Kondo, D. B. Nedwell, K. J. Purdy and Q. Silva, Geomicrobiol. J., 2004, 21, 145–157 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |