Theoretical studies on the dynamics of DNA fragment translocation through multilayer graphene nanopores†
Abstract
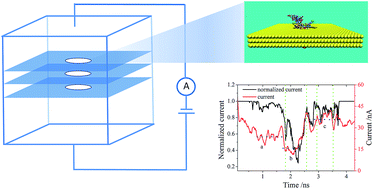
Motivated by several potential advantages over common sequencing technologies, solid-state nanopores, in particular graphene nanopores, have recently been extensively explored as biosensor materials for DNA sequencing. Studies carried out on monolayer graphene nanopores aiming at single-base resolution have recently been extended to multilayer graphene (MLG) films, indicating that MLG nanopores are superior to their monolayer counterparts for DNA sequencing. However, the underlying dynamics and current change in the DNA translocation to thread MLG nanopores remain poorly understood. In this paper, we report a molecular dynamics study of DNA passing through graphene nanopores of different layers. We show that the DNA translocation time could be extended by increasing the graphene layers up to a moderate number (7) under a high electric field and that the current in DNA translocation undergoes a stepwise change upon DNA going through an MLG nanopore. A model is built to account for the relationship between the current change and the unoccupied volume of the MLG nanopore. We demonstrate that the dynamics of DNA translocation depends specifically on the interaction of nucleotides with the graphene sheet. Thus, our study indicates that the resolution of DNA detection could be improved by increasing the number of graphene layers in a certain range and by modifying the surface of the graphene nanopores.

 Please wait while we load your content...
Please wait while we load your content...