Probing the origin of opposite ion-pair binding behavior for two new calix[4]pyrrole bis-phosphonate receptors†
Teng Wang*a,
Jingjing Liub,
Dongsheng Zhangc and
Hongwei Sund
aCollege of Chemical Engineering, Taishan Medical University, Taian, 271016, P.R. China. E-mail: wangteng@mail.nankai.edu.cn
bCollege of Chemistry and Chemical Engineering, Taishan University, Taian, 271021, P.R. China
cCollege of Radiation, Taishan Medical University, Taian, 271016, P.R. China
dCollege of Chemistry, Nankai University, Tianjin, 300071, P.R. China
First published on 8th September 2014
Abstract
A new family of aryl-extended calix[4]pyrroles with two phoshonate groups can act as ion-pair receptors for guest alkylammonium/phosphonium chloride salts, and the ion-pair binding mode and binding affinity of these hosts are different. In the present contribution, the origin of opposite ion-pair binding behavior for two typical hosts ii and oo towards primary ammonium chloride salt and quaternary phosphonium chloride salt was investigated using quantum mechanics (QM) calculations and a new noncovalent weak interaction analysis method. Two types of arrangements – separated and contact – were taken into account for each complex. The binding energy suggests that contact and separated arrangements are the favorable binding modes for receptor ii and oo respectively. Furthermore, the primary alkylammonium chloride salt binds stronger to ii than to oo, while the quaternary phosphonium chloride salt binds stronger to oo than to ii, in agreement with the experiment. Moreover, geometry, charge transfer based on natural bond orbital (NBO) and noncovalent weak interactions analysis suggest that the hydrogen bonding and cation–π interactions play a critical role in ion-pair recognition of ii and oo respectively due to the different orientation of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups. This work further unveils the mechanism of ion-pair recognition by new calix[4]pyrrole bis-phosphonate receptors, while opening exciting perspectives for the design of stronger and more efficient calix[4]pyrrole-based ion-pair receptors.
O groups. This work further unveils the mechanism of ion-pair recognition by new calix[4]pyrrole bis-phosphonate receptors, while opening exciting perspectives for the design of stronger and more efficient calix[4]pyrrole-based ion-pair receptors.
Introduction
Ion-pair recognition is a fast growing research field in supramolecular chemistry.1–6 In comparison with monotopic or single ion receptors that are designed to bind a cation or an anion alone, ditopic or ion-pair receptors with two disparate binding sites can simultaneously bind both cationic and anionic guest species. They have advantages in terms of substrate affinity and selectivity over monotopic receptors. Therefore, ion-pair receptors have potential use in salt solubilization,7–9 ion extraction,10–16 trans-membrane ion transport,17–22 ion sensing,23–27 and as logic gates.28–30Calix[4]pyrroles such as meso-octamethylcalix[4]pyrrole (Fig. 1A) are macrocyclic molecules consisting of four pyrrole rings linked via sp3 hybridized carbon atoms in the 2- and 5- positions. They have received significant attention recently due to their ability to bind anions and ion-pair salts.4,6,31–34 Calix[4]pyrrole is generally conformationally flexible and has four typical conformations.31,35,36 It adopts a 1,3-alternate conformation (Fig. 1B) in the absence of a bound anion. On the other hand, with the addition of anionic guests, it adopts the cone conformation (Fig. 1C and D). Then, in cone conformation, the calix[4]pyrrole provides an electron-rich aromatic cavity that serves as a receptor for cation. In addition, it can mainly bind cations and anions in two different modes, defined as close-contact (contact for short, Fig. 1C) and ion-separated (separated for short, Fig. 1D), respectively. In contact arrangement, the anion and cation are held in close proximity. While in separated arrangement, they are bound relatively far from one another.
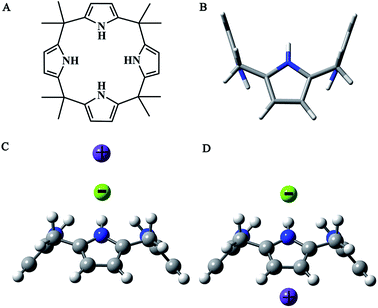 | ||
| Fig. 1 Structure of meso-octamethylcalix[4]pyrrole. (A) Structural formula (B) 1,3-alternate conformation (C) close-contact binding mode (D) ion-separated binding mode. | ||
Considerable effort was devoted to developing calix[4]pyrrole and its derivatives as ion-pair receptors.22,24,26,37,38 Recently, Dalcanale and Ballester et al.39 have reported a new family of aryl-extended calix[4]pyrroles (Fig. 2A) with two phoshonate groups. According to the relative spatial orientation of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups with respect to the aromatic cavity, they were named as ii (Fig. 2B), io, and oo (Fig. 2C) respectively. They can act as ion-pair receptors for alkylammonium/phosphonium (primary and quaternary) chloride salts in dichloromethane (DCM) solution and in the solid state. The ion-pair binding mode and binding affinity are strongly modulated by the spatial orientation of the P
O groups with respect to the aromatic cavity, they were named as ii (Fig. 2B), io, and oo (Fig. 2C) respectively. They can act as ion-pair receptors for alkylammonium/phosphonium (primary and quaternary) chloride salts in dichloromethane (DCM) solution and in the solid state. The ion-pair binding mode and binding affinity are strongly modulated by the spatial orientation of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups. For the ii host, contact arrangement is the favorable binding mode and it binds primary alkylammonium chloride salt most tightly. On the contrary, for the oo host, separated arrangement is the favorable binding mode and it forms the most stable complexes with quaternary alkylphosphonium chloride salt.
O groups. For the ii host, contact arrangement is the favorable binding mode and it binds primary alkylammonium chloride salt most tightly. On the contrary, for the oo host, separated arrangement is the favorable binding mode and it forms the most stable complexes with quaternary alkylphosphonium chloride salt.
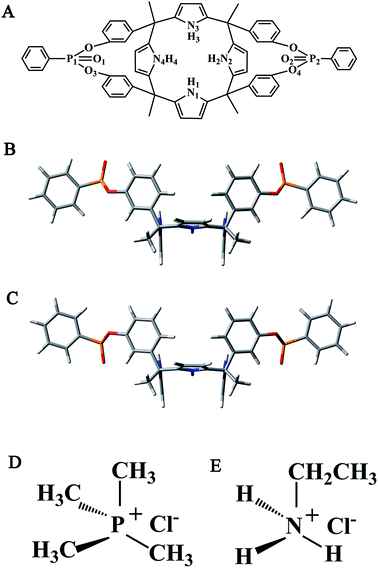 | ||
| Fig. 2 Calix[4]pyrrole bis-phosphonate hosts and alkylphosphonium/ammonium chloride salts. (A) Structural formula with selected atom labels (B) ii (C) oo (D) TMPCl (E) EAMCl. | ||
Most recently, to explore the mechanism of ion-pair recognition by these new receptors, the ion-pair binding mode and binding affinity of the ii host towards quaternary alkylphosphonium chloride salt (tetramethylphosphonium chloride TMPCl, Fig. 2D) and primary alkylammonium chloride salt (ethylammonium chloride EAMCl, Fig. 2E) had been studied using quantum mechanics (QM) calculations.40 We found that contact arrangement was the favorable binding mode for ii both in the gas-phase and in solution, in agreement with the experiment. Moreover, we also predicted that it preferred to bind primary ammonium chloride salt rather than quaternary phosphonium chloride salt. Furthermore, the main driving force responsible for the ion-pair recognition of ii had been revealed and hydrogen bonding interactions played the dominant role in this process.
However, as only ii host was considered to study ion-pair recognition in our previous work, the reason why there are differences in ion-pair binding behavior among these receptors is also not clear. In addition, traditional density functional theory (DFT) B3LYP method41–43 was employed in previous paper and it may give inaccurate results in describing noncovalent interactions.
In the present work, QM calculations have been performed to study the origin of opposite ion-pair binding behavior including binding mode and binding affinity for two new calix[4]pyrrole bis-phosphonate receptors ii and oo both in the gas-phase and in DCM solution. To obtain more accurate noncovalent interactions, the new ωB97X-D44,45 DFT-functional with dispersion corrections was employed. Thus, the ii host and its complexes investigated in our previous work were recalculated using this method. The ion-pair complexes of ii and oo were constructed and fully optimized. The geometrical feature, the binding affinity, and the charge transfer have been discussed in detail. Moreover, the type and strength of noncovalent weak interactions have been explored to unravel the mechanism responsible for the different ion-pair binding behavior of these two receptors.
Methods
The initial geometry of the hosts were taken from the published crystal structure,39 and then the chloride anion and guest cations were introduced to build host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl− and host
Cl− and host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl−
Cl−![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cation complexes. Two types of arrangements – contact and separated were considered for each guest cation. The geometry of the complexes was fully optimized in the gas-phase using DFT at the ωB97X-D/6-31G* level. The harmonic vibrational frequencies were also calculated at the same level of theory in order to confirm whether the obtained geometries corresponded to the energetic minima. Only a few very small imaginary frequencies (less than 50i cm−1) were found for individual structures. This is because the numerical integration procedures used in DFT methods have significant limitations.46 Thus, these low magnitude imaginary vibrational frequencies should imply that there is a genuine minimum of energy identical to or very close to the stationary point, and we do not follow the imaginary eigenvector in search of another minimum.
cation complexes. Two types of arrangements – contact and separated were considered for each guest cation. The geometry of the complexes was fully optimized in the gas-phase using DFT at the ωB97X-D/6-31G* level. The harmonic vibrational frequencies were also calculated at the same level of theory in order to confirm whether the obtained geometries corresponded to the energetic minima. Only a few very small imaginary frequencies (less than 50i cm−1) were found for individual structures. This is because the numerical integration procedures used in DFT methods have significant limitations.46 Thus, these low magnitude imaginary vibrational frequencies should imply that there is a genuine minimum of energy identical to or very close to the stationary point, and we do not follow the imaginary eigenvector in search of another minimum.
To evaluate the binding energy in ion-pair recognition, the complexation reaction steps was designed, as depicted in Fig. 3. The E1deform and E2deform are geometric deformation energy in binding chloride anion and cation respectively. The E1interaction and E2interaction are interaction energy between chloride anion and host and between cation and host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl− complex respectively. Thus, the chloride anion binding energy E1binding and the cation binding energy E2binding, can be computed from the equation as follows respectively:
Cl− complex respectively. Thus, the chloride anion binding energy E1binding and the cation binding energy E2binding, can be computed from the equation as follows respectively:
| E1binding = E1deform + E1interaction | (1) |
| E2binding = E2deform + E2interaction | (2) |
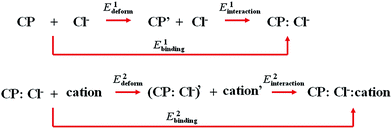 | ||
| Fig. 3 Complexation reaction steps used to evaluate the binding energy in ion-pair recognition. CP represents the ii or oo host. | ||
Therefore, the sum of E1binding and E2binding yields the total ion-pair binding energy Ebinding. On the basis of optimized structures, the ωB97X-D method with 6-31++g** basis set was employed to calculate the binding energy. Moreover, the basis sets superposition error (BSSE) correction was taken into account to obtain an accurate interaction energy by using the counterpoise correction method.47
Based on the optimized structures in the gas-phase, the self-consistent reaction field (SCRF) method was chosen to model the DCM solvent environment to study solvation effects. In the SCRF calculations, Tomasi's polarized continuum model (PCM)48 was used to calculate the binding energy using ωB97X-D method with 6-31++g** basis set.
To investigate the charge transfer in ion-pair recognition, the natural bond orbital (NBO)49 analysis was implemented for the optimized structures by using the ωB97X-D method with 6-31++g** basis set.
To analyze and visualize the noncovalent weak interactions between the host and guest, the electron density (ρ(r)) function, sign(λ2(r))ρ(r) function (sign(λ2(r)) means the sign of the second largest eigenvalue of electron density Hessian matrix at position r), and the reduced density gradient 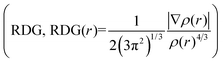 function were calculated respectively. According to Yang's method,50 noncovalent weak interactions are highly nonlocal and manifest in real space as low-gradient isosurfaces with low densities. The sign(λ2(r)) is used to give the type of interaction, and its strength can be derived from the density on the weak interaction surface. On the basis of optimized structures, utilizing different colors to represent sign(λ2(r))ρ(r) function value and map them onto RDG isosurfaces by Multiwfn 3.051 and VMD 1.9,52 the type and the strength of weak interaction can be revealed visually.
function were calculated respectively. According to Yang's method,50 noncovalent weak interactions are highly nonlocal and manifest in real space as low-gradient isosurfaces with low densities. The sign(λ2(r)) is used to give the type of interaction, and its strength can be derived from the density on the weak interaction surface. On the basis of optimized structures, utilizing different colors to represent sign(λ2(r))ρ(r) function value and map them onto RDG isosurfaces by Multiwfn 3.051 and VMD 1.9,52 the type and the strength of weak interaction can be revealed visually.
All calculations were performed using the Gaussian 09 program.53
Results and discussions
Geometry
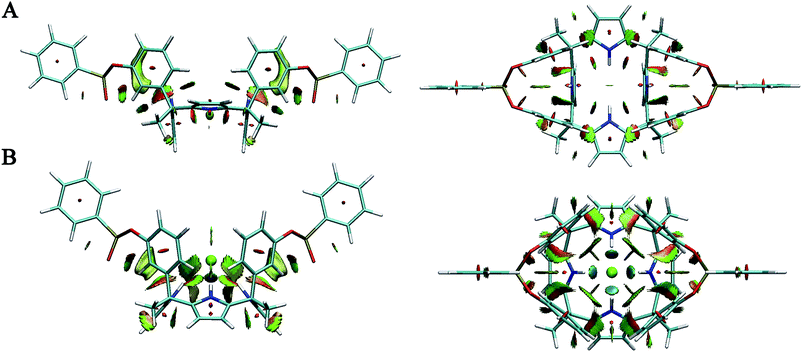
The optimized structures of ii and ii complexes are shown in Fig. S1 (in the ESI†), and the geometrical parameters are reported in Table S1 (in the ESI†). First of all, the symmetry of the corresponding molecules optimized by ωB97X-D method are the same with those by B3LYP method as used in our previous work. Second, quantitatively very similar geometrical parameters including the amount and strength of hydrogen bonds are obtained from the two different methods. Therefore, the structural features calculated from the two methods are basically similar.For oo and oo complexes, the optimized structures are represented in Fig. 4 and 5, and the corresponding structural data are gathered in Table 1. As shown in Fig. 4, the calix[4]pyrrole fragment transforms from a 1,3-alternate conformation in the free host to a cone conformation when binding chloride anion through N–H⋯Cl hydrogen bonds. In addition, the distances P1⋯P2 and O1⋯O2 are shorter in oo–Cl than that in oo, showing that the benzene rings are induced to come close to the chloride anion through anion–π interactions. These interactions can be confirmed in the following weak interaction analysis.
 | ||
| Fig. 5 Side and top views of optimized structures of the oo–TMPCl and oo–EAMCl. (A) oo–TMPCl–1 (B) oo–TMPCl–2 (C) oo–EAMCl–1 (D) oo–EAMCl–2. | ||
| Parameter | oo | oo–Cl | oo–TMPCl–1 | oo–TMPCl–2 | oo–EAMCl–1 | oo–EAMCl–2 |
|---|---|---|---|---|---|---|
| Symmetry | C2v | C2v | C1 | C1 | C1 | C1 |
| N1–H1 | 1.01 | 1.03 | 1.02 | 1.03 | 1.02 | 1.03 |
| N2–H2 | 1.01 | 1.02 | 1.02 | 1.03 | 1.01 | 1.03 |
| N3–H3 | 1.01 | 1.03 | 1.02 | 1.03 | 1.02 | 1.03 |
| N4–H4 | 1.01 | 1.02 | 1.02 | 1.03 | 1.02 | 1.03 |
| H1⋯Cl | — | 2.22 | 2.27 | 2.16 | 2.24 | 2.24 |
| H2⋯Cl | — | 2.24 | 2.25 | 2.21 | 2.24 | 2.24 |
| H3⋯Cl | — | 2.22 | 2.23 | 2.23 | 2.29 | 2.18 |
| H4⋯Cl | — | 2.24 | 2.25 | 2.21 | 2.29 | 2.22 |
| ∠N1–H1⋯Cl | — | 178.83 | 175.23 | 173.30 | 173.00 | 156.05 |
| ∠N2–H2⋯Cl | — | 168.01 | 173.16 | 165.67 | 171.15 | 157.58 |
| ∠N3–H3⋯Cl | — | 178.83 | 178.97 | 166.51 | 174.12 | 169.23 |
| ∠N4–H4⋯Cl | — | 168.01 | 173.33 | 165.67 | 178.53 | 162.46 |
| P3⋯Cl | — | — | 3.51 | 6.09 | — | — |
| P1⋯P2 | 14.03 | 11.61 | 11.32 | 11.45 | 10.87 | 11.32 |
| O1⋯O2 | 13.65 | 12.64 | 12.36 | 12.44 | 12.11 | 12.32 |
| O3⋯Ha | — | — | 2.48 | — | — | — |
| O4⋯Hb | — | — | 2.50 | — | — | — |
| ∠C1–Ha⋯O3 | — | — | 134.29 | — | — | — |
| ∠C2–Hb⋯O4 | — | — | 132.82 | — | — | — |
| N5⋯Cl | — | — | — | — | 2.93 | 4.11 |
| H5⋯Cl | — | — | — | — | 1.94 | — |
| O3⋯H6 | — | — | — | — | 2.68 | — |
| O3⋯H7 | — | — | — | — | 2.60 | — |
| ∠N5–H5⋯Cl | — | — | — | — | 155.33 | — |
| ∠N5–H6⋯O3 | — | — | — | — | 97.17 | — |
| ∠N5–H7⋯O3 | — | — | — | — | 101.30 | — |
| H5⋯N4 | — | — | — | — | — | 2.02 |
| H6⋯N1 | — | — | — | — | — | 2.55 |
| H7⋯N2 | — | — | — | — | — | 1.98 |
| ∠N5–H5⋯N4 | — | — | — | — | — | 159.55 |
| ∠N5–H6⋯N1 | — | — | — | — | — | 105.05 |
| ∠N5–H7⋯N2 | — | — | — | — | — | 179.62 |
For contact arrangement oo–TMPCl–1, two weak C–H⋯O hydrogen bonds are formed between phosphonate groups and methyl groups. Because the phosphonate groups directed outwardly, the TMP cation forms hydrogen bonds with oxygen atoms in P–O single bonds while it forms hydrogen bonds with oxygen atoms in P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O double bonds in ii–TMPCl–1a and ii–TMPCl–1b. Moreover, the O⋯H distances and C–H⋯O angles are longer and smaller in oo–TMPCl–1 than that in ii–TMPCl–1a (both are 2.09 Å and 156.14°) and ii–TMPCl–1b (both are 2.17 Å and 134.38°) respectively, suggesting that the C–H⋯O hydrogen bonds in the former are weaker than that in the latter. For separated arrangement oo–TMPCl–2, no hydrogen bonds can be found between host and TMP cation.
O double bonds in ii–TMPCl–1a and ii–TMPCl–1b. Moreover, the O⋯H distances and C–H⋯O angles are longer and smaller in oo–TMPCl–1 than that in ii–TMPCl–1a (both are 2.09 Å and 156.14°) and ii–TMPCl–1b (both are 2.17 Å and 134.38°) respectively, suggesting that the C–H⋯O hydrogen bonds in the former are weaker than that in the latter. For separated arrangement oo–TMPCl–2, no hydrogen bonds can be found between host and TMP cation.
For contact arrangement oo–EAMCl–1, three hydrogen bonds are established, viz. N5–H5⋯Cl, N5–H6⋯O3, and N5–H7⋯O3, and these hydrogen bonds are relatively weak from the geometric viewpoint. Hydrogen bond N5–H5⋯Cl is stronger than hydrogen bonds N5–H6⋯O3 and N5–H7⋯O3. As the phosphonate groups are pointing outwardly, the EAM cation can only form hydrogen bonds with one phosphonate group while it forms hydrogen bonds with two phosphonate groups in ii–EAMCl–1. In addition, the O⋯H distances and N–H⋯O angles are longer and smaller in oo–EAMCl–1 than that in ii–EAMCl–1 (both are 1.88 Å and 143.72°) respectively, suggesting that the N–H⋯O hydrogen bonds in the former are weaker than that in the latter. Although the N–H⋯Cl hydrogen bond in oo–EAMCl–1 is slightly stronger than that in ii–EAMCl–1 (the H⋯Cl distance and N–H⋯Cl angle are 2.08 Å and 147.13° respectively), the total hydrogen bonding interactions in the former are weaker than that in the latter. For separated arrangement oo–EAMCl–2, there are also three hydrogen bonds are formed, viz. N5–H5⋯N4, N5–H6⋯N1, and N5–H7⋯N2. Furthermore, the distance H7⋯N2 is the shortest and the angle ∠N5–H7⋯N2 is the largest among the three hydrogen bonds, indicating that hydrogen bond N5–H7⋯N2 is stronger than hydrogen bonds N5–H5⋯N4 and N5–H6⋯N1.
Weak interaction
To investigate the weak interaction visually, the gradient isosurfaces of RDG for oo host and its complexes are delineated in Fig. 6 and 7. The surfaces are colored on a blue-green-red scale according to values of sign(λ2(r))ρ(r) over the range from −0.04 to 0.02 au. Blue indicates strong attraction, red indicates strong repulsion, and green indicates weak attractive interactions.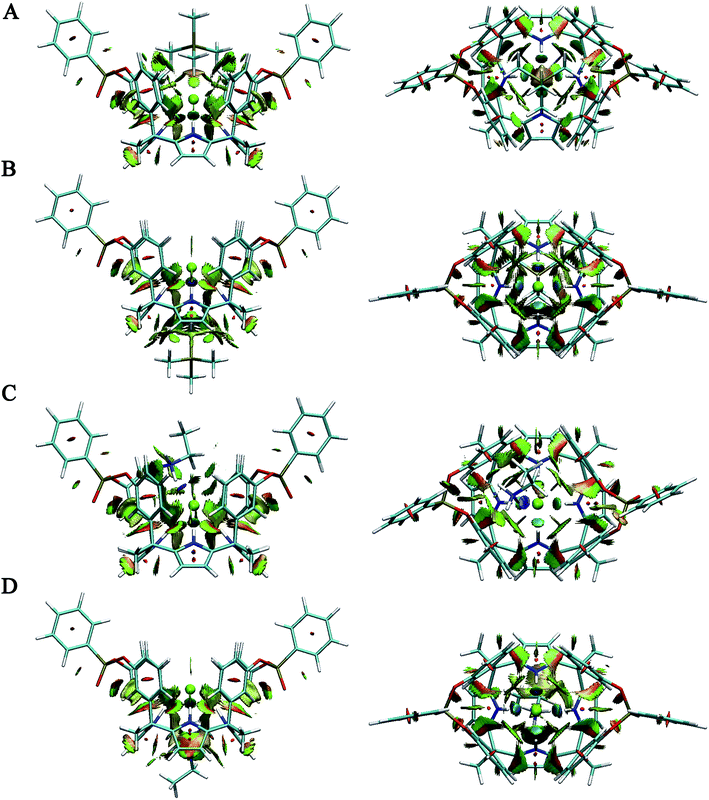 | ||
| Fig. 7 Side and top views of gradient isosurfaces (RDG = 0.5 au) for (A) oo–TMPCl–1 (B) oo–TMPCl–2 (C) oo–EAMCl–1 (D) oo–EAMCl–2. | ||
Similar to the ii host, there is steric repulsion (orange regions of the isosurface) between each pyrrole ring and its neighboring benzene rings and pyrrole rings in oo host (Fig. 6A) and that is the reason for energy barrier of conformational transition from 1,3-alternate to cone conformation. For oo–Cl complex (Fig. 6B), light blue regions of the isosurface can be clearly detected and these indicate N–H⋯Cl hydrogen bonding interactions. In addition, there are green regions between the chloride anion and four benzene rings and can be identified as anion–π interactions.
For oo–TMPCl–1 (Fig. 7A), two weak C–H⋯O hydrogen bonds between methyls groups of TMP and phosphonate groups of oo can be found. Moreover, there are green areas between methyls groups and benzene rings, suggesting that there are cation–π interactions between TMP and oo. However, these green areas are very small and showing that the cation–π interactions are weak. In addition, van der Waals interactions between TMP and chloride anion can also be revealed. For oo–TMPCl–2 (Fig. 7B), the strong cation–π interactions between TMP and pyrrole rings of oo can be detected by large green areas. Furthermore, the charge–charge interactions between cations and anions for separated arrangement are stronger than those for contact arrangement according to the following NBO analysis. Although the charge–charge interactions are strong, these interactions cannot be displayed in gradient isosurfaces because this gradient isosurface is extremely small. Additionally, due to the large distance between TMP cation and chloride anion, the van der Waals interactions between them are very weak.
For oo–EAMCl–1 (Fig. 7C), N–H⋯O and N–H⋯Cl hydrogen bonds between EAM and oo can be identified. Among them, N–H⋯Cl hydrogen bond is strongest and can be located by blue area, in agreement with the geometry analysis above. In addition, there are weak cation–π and van der Waals interactions between EAM and benzene rings and between EAM and chloride anion respectively. For oo–EAMCl–2 (Fig. 7D), three N–H⋯N hydrogen bonds can be detected, and hydrogen bond N5–H7⋯N2 is stronger than hydrogen bonds N5–H5⋯N4 and N5–H6⋯N1, in agreement with the geometry analysis above. Moreover, there are large green areas between EAM and pyrrole rings, and they can be identified as strong cation–π interactions. In addition, the charge–charge interactions between cations and anions for separated arrangement are still stronger than those for contact arrangement and also cannot be displayed. The van der Waals interactions between EAM cation and chloride anion are also weak due to the large distance.
Charge transfer
To study the charge transfer in ion-pair recognition, charges of ions and hosts in oo complexes based on NBO analysis is reported in Table 2. For the ion-pair complexes of ii, the behavior of charge transfer is basically the same with that of B3LYP results (see Table S2†).| oo–Cl | oo–TMPCl–1 | oo–TMPCl–2 | oo–EAMCl–1 | oo–EAMCl–2 | |
|---|---|---|---|---|---|
| Cl | −0.766 | −0.800 | −0.740 | −0.733 | −0.749 |
| TMP | — | 0.951 | 0.958 | — | — |
| EAM | — | — | — | 0.858 | 0.919 |
| oo | −0.234 | −0.151 | −0.218 | −0.125 | −0.170 |
| Dipole moment | 9.519 | 18.664 | 8.244 | 16.161 | 1.351 |
For oo–Cl, electron transfer from chloride anion to the oo host through the N–H⋯Cl hydrogen bonding interactions. For oo–TMPCl–1, the charges of oo and chloride anion are less and more negative than that in oo–Cl respectively. The electron transfer from oo to the TMP through C–H⋯O hydrogen bonding interactions when binding this cation. On the other hand, the hydrogen bonds bring benzene rings of oo closer, and then some electron transfer from benzene rings of oo to the chloride anion due to the enhanced anion–π interactions. For oo–TMPCl–2, the negative charges of oo and chloride anion are both reduced compared to that in oo–Cl, indicating that the electron transfer from oo and chloride anion to TMP cation when binding it. Moreover, this charge transfer is just carried out by direct or indirect cation–π interactions. The electron transfer from oo to TMP through direct cation–π interactions. On the other hand, the electron of chloride anion transfer to the oo through N–H⋯Cl hydrogen bonding interactions, and then these electron transfer from oo that can be taken as the media of charge transfer to TMP through cation–π interactions. For oo–EAMCl–1, the electron transfer from oo and chloride anion to EAM cation through N–H⋯O and N–H⋯Cl hydrogen bonding interactions respectively. For oo–EAMCl–2, the electron transfer from oo to EAM cation through N–H⋯N hydrogen bonding and cation–π interactions, and the electron transfer from chloride anion to EAM cation through media oo.
The charge transfer comes from hydrogen bonding and cation–π interactions when binding cations, and the hydrogen bonding interactions hold a high percentage in it. Thus the charge transfer can reflect the strength of hydrogen bonds. The cations accept more negative charge in contact arrangements oo–TMPCl–1 and oo–EAMCl–1 than that in separated arrangements oo–TMPCl–2 and oo–EAMCl–2 respectively. The results show that the hydrogen bonding interactions in contact arrangements are stronger than that in separated arrangements. Furthermore, the change of charge for cation in primary alkylammonium chloride salt complexes is larger than that in quaternary phosphonium chloride salt complexes, suggesting that the hydrogen bonding interactions in the former are stronger than that in the latter.
In addition, the dipole moments of oo complexes are also given in Table 2. For the ion-pair complexes of oo, the dipole moments of separated arrangements complexes are much smaller than that of contact arrangements complexes. This means that the centers of positive and negative charge approach much closer to each other in the former than they are in the latter. Therefore, the charge–charge interactions between ion-pair in separated arrangements are much stronger than that in contact arrangements. Although the hydrogen bonding interactions in separated arrangements are weaker than contact arrangements for oo complexes, the strong cation–π and charge–charge interactions make the former arrangements more energetically favorable.
Binding energy
To study the binding affinity of receptor ii and oo towards ion-pairs TMPCl and EAMCl, the binding energy in the gas-phase were calculated, as shown in Table 3.| Complex | E1deform | E1interaction | E1interaction (BSSE) | E1binding | E1binding (BSSE) |
|---|---|---|---|---|---|
| ii–Cl | 10.70 | −57.58 | −56.70 | −46.88 | −46.00 |
| E2deform | E2interaction | E2interaction (BSSE) | E2binding | E2binding (BSSE) | Ebinding | Ebinding (BSSE) | |
|---|---|---|---|---|---|---|---|
| ii–TMPCl–1a | 4.65 | −98.94 | −96.50 | −94.29 | −91.85 | −141.17 | −137.85 |
| ii–TMPCl–1b | 4.17 | −100.32 | −97.79 | −96.15 | −93.62 | −143.03 | −139.62 |
| ii–TMPCl–2a | 2.02 | −93.05 | −90.88 | −91.03 | −88.86 | −137.91 | −134.86 |
| ii–TMPCl–2b | 1.79 | −93.02 | −90.88 | −91.23 | −89.09 | −138.11 | −135.09 |
| ii–EAMCl–1 | 8.75 | −133.98 | −131.43 | −125.23 | −122.68 | −172.11 | −168.68 |
| ii–EAMCl–2 | 4.39 | −118.34 | −116.23 | −113.95 | −111.84 | −160.83 | −157.84 |
| Complex | E1deform | E1interaction | E1interaction (BSSE) | E1binding | E1binding (BSSE) |
|---|---|---|---|---|---|
| oo–Cl | 11.77 | −57.83 | −57.02 | −46.06 | −45.25 |
| E2deform | E2interaction | E2interaction (BSSE) | E2binding | E2binding (BSSE) | Ebinding | Ebinding (BSSE) | |
|---|---|---|---|---|---|---|---|
| oo–TMPCl–1 | 3.57 | −90.79 | −88.36 | −87.22 | −84.79 | −133.28 | −130.04 |
| oo–TMPCl–2 | 1.30 | −95.91 | −93.71 | −94.61 | −92.41 | −140.67 | −137.66 |
| oo–EAMCl–1 | 2.15 | −107.54 | −105.90 | −105.39 | −103.75 | −151.45 | −149.00 |
| oo–EAMCl–2 | 3.50 | −121.35 | −119.19 | −117.85 | −115.69 | −163.91 | −160.94 |
For receptor ii, contact arrangements ii–TMPCl–1a, ii–TMPCl–1b and ii–EAMCl–1 are more favorable than separated arrangements ii–TMPCl–2a, ii–TMPCl–2b and ii–EAMCl–2 respectively, in agreement with the results of experiment and B3LYP method. Furthermore, although pairwise competitive binding experiments were not conclusive for rating the binding affinity of ii with respect to TMPCl and EAMCl, the computational results indicate that it prefers to bind primary ammonium chloride salt rather than quaternary phosphonium chloride salt, also in agreement with the results of B3LYP method. In addition, the binding energy calculated from ωB97X-D method are more favorable than that from B3LYP method for the corresponding complexation reaction step, and this is because the former with dispersion corrections can account for dispersion binding. Although the absolute binding energy calculated from the two methods are different, the relative binding affinity are same. The similar case also can be found in the following liquid-phase calculation.
For receptor oo, the binding energy of separated arrangements oo–TMPCl–2 and oo–EAMCl–2 are ca. 7.6 kcal mol−1 and 12 kcal mol−1 lower than that of contact arrangements oo–TMPCl–1 and oo–EAMCl–1 respectively. It is suggested that separated arrangement is the favorable binding mode for receptor oo, in agreement with the experiment. The reason can be explained in the following way. For oo–TMPCl complexes, only two weak C–H⋯O hydrogen bonds are formed in contact arrangement, while there are strong cation–π interactions between cation and pyrrole rings in separated arrangement. For oo–EAMCl complexes, although the hydrogen bond strength of one N–H⋯Cl and two N–H⋯O in contact arrangement are stronger than that of three N–H⋯N in separated arrangement, the cation–π interactions make a favorable contribution to the binding energy for the latter. In addition, the charge–charge interactions between ion-pair in separated arrangements are much stronger than that in contact arrangements as mentioned in dipole moments analysis. Therefore, oo prefers to separated arrangement rather than contact arrangement.
For TMPCl, the calculated binding energy of ii are lower than that of oo for each favorable arrangement, showing that ii binds TMPCl more strongly than oo in the gas-phase. However, according to experimental results, the order of binding affinity for the receptors towards TMPCl is oo ≈ 4 ii in DCM solvent. To study the solvation effects, the binding energy of contact arrangement for ii and two arrangements for oo were calculated in the DCM solvent and presented in Table 4. The results show that oo binds TMPCl more strongly than ii in the DCM solvent, in agreement with the experimental results. Firstly, the binding energy of oo–Cl is lower than that of ii–Cl, indicating that the binding affinity of chloride anion to the oo is greater than that of it to ii. This can be ascribed to the orientation of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups. The two oxygen atoms of the P
O groups. The two oxygen atoms of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups are pointing inwardly with respect to the deep cavity in ii and there will be repulsive electrostatic interactions between the inwardly directed oxygen atoms of ii and the chloride anion. While the two oxygen atoms of P
O groups are pointing inwardly with respect to the deep cavity in ii and there will be repulsive electrostatic interactions between the inwardly directed oxygen atoms of ii and the chloride anion. While the two oxygen atoms of P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in oo directed outwardly and there are no repulsive electrostatic interactions between oxygen atoms and the chloride anion. Therefore, although the E1deform of oo–Cl is less favorable than that of ii–Cl, the E1interaction of the former is ca. 3 kcal mol−1 lower than that of the latter. Then, the E2interaction of oo–TMPCl–2 is ca. 2.5 kcal mol−1 lower than that of ii–TMPCl–1b, suggesting that oo–Cl binds TMP cation more strongly than ii–Cl. Although there are C–H⋯O hydrogen bonds between cation and phosphonate groups in ii–TMPCl–1b, they are very weak. While there are strong cation–π interactions between cation and pyrrole rings in oo–TMPCl–2. Therefore, oo binds TMPCl more strongly than ii and cation–π interactions play the dominant role in this process. In addition, the solvation effects have an important influence on the ion-pair recognition.
O groups in oo directed outwardly and there are no repulsive electrostatic interactions between oxygen atoms and the chloride anion. Therefore, although the E1deform of oo–Cl is less favorable than that of ii–Cl, the E1interaction of the former is ca. 3 kcal mol−1 lower than that of the latter. Then, the E2interaction of oo–TMPCl–2 is ca. 2.5 kcal mol−1 lower than that of ii–TMPCl–1b, suggesting that oo–Cl binds TMP cation more strongly than ii–Cl. Although there are C–H⋯O hydrogen bonds between cation and phosphonate groups in ii–TMPCl–1b, they are very weak. While there are strong cation–π interactions between cation and pyrrole rings in oo–TMPCl–2. Therefore, oo binds TMPCl more strongly than ii and cation–π interactions play the dominant role in this process. In addition, the solvation effects have an important influence on the ion-pair recognition.
| Complex | E1deform | E1interaction | E1binding |
|---|---|---|---|
| ii–Cl | 5.60 | −21.87 | −16.27 |
| E2deform | E2interaction | E2binding | Ebinding | Ka,exp | ||
|---|---|---|---|---|---|---|
| 1H NMR titration experiments | Isothermal titration calorimetry (ITC) experiments | |||||
| ii–TMPCl–1a | 0.70 | −25.41 | −24.71 | −40.98 | >104 | 2 ± 0.5 × 105 |
| ii–TMPCl–1b | −0.43 | −25.51 | −25.94 | −42.21 | ||
| ii–EAMCl–1 | 4.31 | −43.52 | −39.21 | −55.48 | >104 | — |
| Complex | E1deform | E1interaction | E1binding |
|---|---|---|---|
| oo–Cl | 7.99 | −24.66 | −16.67 |
| E2deform | E2interaction | E2binding | Ebinding | Ka,exp | ||
|---|---|---|---|---|---|---|
| 1H NMR titration experiments | Isothermal titration calorimetry (ITC) experiments | |||||
| oo–TMPCl–1 | 0.67 | −22.61 | −21.94 | −38.61 | >105 | 8 ± 1 × 105 |
| oo–TMPCl–2 | −0.71 | −28.02 | −28.73 | −45.40 | ||
| oo–EAMCl–1 | 0.50 | −27.11 | −26.61 | −43.28 | 2 ± 1 × 103 | — |
| oo–EAMCl–2 | 1.52 | −35.47 | −33.95 | −50.62 | ||
For EAMCl, the calculated binding energy of ii are lower than that of oo for each favorable arrangement both in the gas-phase and DCM solvent, showing that ii binds EAMCl more strongly than oo. This is in agreement with the experimental results. The main reason is that there are two strong N–H⋯O hydrogen bonds and one strong N–H⋯Cl hydrogen bond in ii–EAMCl–1, and there are only three weak N–H⋯N hydrogen bonds in oo–EAMCl–2. Although there are strong cation–π interactions between cation and pyrrole rings in oo–EAMCl–2, it is weaker than the strong hydrogen bonding interactions in ii–EAMCl–1. Thus, the different binding affinity for the receptors towards EAMCl can be mainly ascribed to the hydrogen bonding interactions.
In addition, the calculation results show that oo binds EAMCl more strongly than TMPCl both in the gas-phase and DCM solvent. For oo–EAMCl–2, the favorable arrangement of oo–EAMCl complex, there are N–H⋯N hydrogen bonds and cation–π interactions between cation and host. While there are only cation–π interactions in favorable arrangement of oo–TMPCl complex – oo–TMPCl–2. Therefore, it seems that oo prefers to bind EAMCl rather than TMPCl. However, it is in contradiction with the experimental findings. The reason for this might be that the cation–π interactions in oo–TMPCl–2 are stronger than hydrogen bonding and cation–π interactions in oo–EAMCl–2, and our calculation method cannot reflect this phenomenon very well.
Conclusions
The origin of different ion-pair binding behavior for new calix[4]pyrrole bis-phosphonate receptors ii and oo has been explored by QM calculations. The calculated binding energy of the ion-pair recognition suggests that contact and separated arrangements are the favorable binding modes for receptor ii and oo respectively. Furthermore, the primary alkylammonium chloride salt EAMCl binds stronger to ii than to oo, while quaternary phosphonium chloride salt TMPCl binds stronger to oo than to ii, in agreement with the experiment. Moreover, the computational results show that ii prefers to bind EAMCl rather than TMPCl, despite the experiments cannot give the binding affinity of ii with respect to these two guests. In addition, the calculation results also suggest that oo binds EAMCl more strongly than TMPCl, however, in contradiction with the experiment. The ion-pair binding behavior for ii and oo can be summed in Table 5.| ii | oo | |
|---|---|---|
| Critical interactions | Hydrogen bonding | Cation–π |
| Favorable binding modes | Contact | Separated |
| EAMCl binding affinity | ii > oo | |
| TMPCl binding affinity | oo > ii | |
| Favorable binding guests | EAMCl | TMPCl |
Hydrogen bonding and cation–π interactions play the critical role in ion-pair recognition of ii and oo respectively due to the different orientation of the P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups. Concretely, the hydrogen bonding interactions between ii and guests in contact arrangements are stronger than that in separated arrangements, and cation–π interactions between oo and guests in separated arrangements are stronger than that in contact arrangements. Moreover, the charge–charge interactions between cations and anions in oo complexes for separated arrangements are stronger than those for contact arrangements due to the smaller dipole moment. Therefore, ii and oo prefer to contact and separated arrangements respectively. The cation–π interactions between oo and TMPCl in separated arrangement are stronger than hydrogen bonding interactions between ii and TMPCl in contact arrangement, and the hydrogen bonding interactions between ii and EAMCl in contact arrangement are stronger than cation–π interactions between oo and EAMCl in separated arrangement. This is the reason for different ion-pair binding affinity for ii and oo. In addition, the hydrogen bonding interactions between ii and EAMCl are stronger than that between ii and TMPCl, and the cation–π interactions between oo and TMPCl are stronger than that between oo and EAMCl. This may explain why ii and oo prefer to bind EAMCl and TMPCl respectively.
O groups. Concretely, the hydrogen bonding interactions between ii and guests in contact arrangements are stronger than that in separated arrangements, and cation–π interactions between oo and guests in separated arrangements are stronger than that in contact arrangements. Moreover, the charge–charge interactions between cations and anions in oo complexes for separated arrangements are stronger than those for contact arrangements due to the smaller dipole moment. Therefore, ii and oo prefer to contact and separated arrangements respectively. The cation–π interactions between oo and TMPCl in separated arrangement are stronger than hydrogen bonding interactions between ii and TMPCl in contact arrangement, and the hydrogen bonding interactions between ii and EAMCl in contact arrangement are stronger than cation–π interactions between oo and EAMCl in separated arrangement. This is the reason for different ion-pair binding affinity for ii and oo. In addition, the hydrogen bonding interactions between ii and EAMCl are stronger than that between ii and TMPCl, and the cation–π interactions between oo and TMPCl are stronger than that between oo and EAMCl. This may explain why ii and oo prefer to bind EAMCl and TMPCl respectively.
The main reason responsible for the different ion-pair binding mode and binding affinity for new calix[4]pyrrole bis-phosphonate receptors ii and oo has been revealed at the electronic level, providing valuable insights into the potential of these calix[4]pyrrole-based supramolecular systems to act as ion-pair receptors. To better understand the origin of different ion-pair binding behavior, further investigation on the ion-pair recognition of transition receptor io is still needed. Moreover, other computational methods such as molecular dynamics simulations combined with free-energy calculations that may give accurate results in binding affinity and describing solvation effects are also needed, which are beyond the scope of this study.
Acknowledgements
This study was supported by the Research Award Fund for Outstanding Middle-Aged and Young Scientists of Shandong Province, China (no. BS2013YY061), the Natural Sciences Foundation of Shandong Province, China (no. ZR2013BL014), the Taishan Medical University (no. 2012GCC07) and the Taishan University (no. Y-01-2014013).References
- G. J. Kirkovits, J. A. Shriver, P. A. Gale and J. L. Sessler, J. Inclusion Phenom. Macrocyclic Chem., 2001, 41, 69–75 CrossRef CAS.
- P. D. Beer and P. A. Gale, Angew. Chem., Int. Ed., 2001, 40, 486–516 CrossRef CAS.
- B. D. Smith in Macrocyclic Chemistry: Current Trends and Future Perspectives, ed. K. Gloe, Springer, Dordrecht, Netherlands, 2005, pp. 137–151 Search PubMed.
- S. K. Kim and J. L. Sessler, Chem. Soc. Rev., 2010, 39, 3784–3809 RSC.
- A. J. McConnell and P. D. Beer, Angew. Chem., Int. Ed., 2012, 51, 5052–5061 CrossRef CAS PubMed.
- M. Wenzel, J. R. Hiscock and P. A. Gale, Chem. Soc. Rev., 2012, 41, 480–520 RSC.
- P. R. A. Webber and P. D. Beer, Dalton Trans., 2003, 2249–2252 RSC.
- G. Reeske, G. Bradtmoller, M. Schurmann and K. Jurkschat, Chem.–Eur. J., 2007, 13, 10239–10245 CrossRef CAS PubMed.
- S. K. Kim, D. E. Gross, D. G. Cho, V. M. Lynch and J. L. Sessler, J. Org. Chem., 2011, 76, 1005–1012 CrossRef CAS PubMed.
- M. J. Deetz, M. Shang and B. D. Smith, J. Am. Chem. Soc., 2000, 122, 6201–6207 CrossRef CAS.
- J. M. Mahoney, A. M. Beatty and B. D. Smith, Inorg. Chem., 2004, 43, 7617–7621 CrossRef CAS PubMed.
- M. P. Wintergerst, T. G. Levitskaia, B. A. Moyer, J. L. Sessler and L. H. Delmau, J. Am. Chem. Soc., 2008, 130, 4129–4139 CrossRef CAS PubMed.
- A. Aydogan, D. J. Coady, S. K. Kim, A. Akar, C. W. Bielawski, M. Marquez and J. L. Sessler, Angew. Chem., Int. Ed., 2008, 47, 9648–9652 CrossRef CAS PubMed.
- R. S. Forgan, J. E. Davidson, F. P. A. Fabbiani, S. G. Galbraith, D. K. Henderson, S. A. Moggach, S. Parsons, P. A. Tasker and F. J. White, Dalton Trans., 2010, 39, 1763–1770 RSC.
- S. K. Kim, V. M. Lynch, N. J. Young, B. P. Hay, C. H. Lee, J. S. Kim, B. A. Moyer and J. L. Sessler, J. Am. Chem. Soc., 2012, 134, 20837–20843 CrossRef CAS PubMed.
- J. Romanski and P. Piatek, J. Org. Chem., 2013, 78, 4341–4347 CrossRef CAS PubMed.
- L. A. J. Chrisstoffels, F. de Jong, D. N. Reinhoudt, S. Sivelli, L. Gazzola, A. Casnati and R. Ungaro, J. Am. Chem. Soc., 1999, 121, 10142–10151 CrossRef CAS.
- J. M. Mahoney, G. U. Nawaratna, A. M. Beatty, P. J. Duggan and B. D. Smith, Inorg. Chem., 2004, 43, 5902–5907 CrossRef CAS PubMed.
- A. P. Davis, D. N. Sheppardb and B. D. Smith, Chem. Soc. Rev., 2007, 36, 348–357 RSC.
- C. C. Tong, R. Quesada, J. L. Sessler and P. A. Gale, Chem. Commun., 2008, 6321–6323 RSC.
- P. A. Gale, Acc. Chem. Res., 2011, 44, 216–226 CrossRef CAS PubMed.
- I. W. Park, J. Yoo, B. Kim, S. Adhikari, S. K. Kim, Y. Yeon, C. J. E. Haynes, J. L. Sutton, C. C. Tong, V. M. Lynch, J. L. Sessler, P. A. Gale and C. H. Lee, Chem.–Eur. J., 2012, 18, 2514–2523 CrossRef CAS PubMed.
- J. M. Mahoney, A. M. Beatty and B. D. Smith, J. Am. Chem. Soc., 2001, 123, 5847–5848 CrossRef CAS.
- S. K. Kim, J. L. Sessler, D. E. Gross, C. H. Lee, J. S. Kim, V. M. Lynch, L. H. Delmau and B. P. Hay, J. Am. Chem. Soc., 2010, 132, 5827–5836 CrossRef CAS PubMed.
- M. Alfonso, A. Espinosa, A. Tarraga and P. Molina, Org. Lett., 2011, 13, 2078–2081 CrossRef CAS PubMed.
- S. K. Kim, G. I. Vargas-Zuniga, B. P. Hay, N. J. Young, L. H. Delmau, C. Masselin, C. H. Lee, J. S. Kim, V. M. Lynch, B. A. Moyer and J. L. Sessler, J. Am. Chem. Soc., 2012, 134, 1782–1792 CrossRef CAS PubMed.
- S. K. Kim, J. M. Lim, T. Pradhan, H. S. Jung, V. M. Lynch, J. S. Kim, D. Kim and J. L. Sessler, J. Am. Chem. Soc., 2014, 136, 495–505 CrossRef CAS PubMed.
- H. Miyaji, S. R. Collinson, I. Prokes and J. H. R. Tucker, Chem. Commun., 2003, 64–65 RSC.
- J. K. Choi, K. No, E. H. Lee, S. G. Kwon, K. W. Kim and J. S. Kim, Supramol. Chem., 2007, 19, 283–286 CrossRef CAS.
- D. S. Kim, V. M. Lynch, J. S. Park and J. L. Sessler, J. Am. Chem. Soc., 2013, 135, 14889–14894 CrossRef CAS PubMed.
- P. A. Gale, J. L. Sessler and V. Kral, Chem. Commun., 1998, 1–8 RSC.
- P. A. Gale, P. Anzenbacher Jr and J. L. Sessler, Coord. Chem. Rev., 2001, 222, 57–102 CrossRef CAS.
- P. A. Gale, J. L. Sessler, V. Kral and V. Lynch, J. Am. Chem. Soc., 1996, 118, 5140–5141 CrossRef CAS.
- R. Custelcean, L. H. Delmau, B. A. Moyer, J. L. Sessler, W. S. Cho, D. Gross, G. W. Bates, S. J. Brooks, M. E. Light and P. A. Gale, Angew. Chem., Int. Ed., 2005, 44, 2537–2542 CrossRef CAS PubMed.
- W. P. van Hoorn and W. L. Jorgensen, J. Org. Chem., 1999, 64, 7439–7444 CrossRef CAS.
- Y. D. Wu, D. F. Wang and J. L. Sessler, J. Org. Chem., 2001, 66, 3739–3746 CrossRef CAS PubMed.
- J. L. Sessler, S. K. Kim, D. E. Gross, C. H. Lee, J. S. Kim and V. M. Lynch, J. Am. Chem. Soc., 2008, 130, 13162–13166 CrossRef CAS PubMed.
- I. W. Park, J. Yoo, S. Adhikari, J. S. Park, J. L. Sessler and C. H. Lee, Chem.–Eur. J., 2012, 18, 15073–15078 CrossRef CAS PubMed.
- M. Ciardi, F. Tancini, G. Gil-Ramirez, E. C. Escudero Adan, C. Massera, E. Dalcanale and P. Ballester, J. Am. Chem. Soc., 2012, 134, 13121–13132 CrossRef CAS PubMed.
- T. Wang, J. J. Liu, H. W. Sun, L. Chen, J. Dong, L. P. Sun and Y. S. Bi, RSC Adv., 2014, 4, 1864–1873 RSC.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS.
- G. A. Petersson and M. A. Al-Laham, J. Chem. Phys., 1991, 94, 6081–6090 CrossRef CAS PubMed.
- J. D. Chai and M. Head-Gordon, J. Chem. Phys., 2008, 128, 084106 CrossRef PubMed.
- J. D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- B. N. Papas and H. F. Schaefer, J. Mol. Struct.: THEOCHEM, 2006, 768, 175–181 CrossRef CAS PubMed.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553–566 CrossRef CAS.
- S. Miertus, E. Scrocco and J. Tomasi, Chem. Phys., 1981, 55, 117–129 CrossRef CAS.
- A. E. Reed, L. A. Curtiss and F. Weinhold, Chem. Rev., 1988, 88, 899–926 CrossRef CAS.
- E. R. Johnson, S. Keinan, P. Mori-Sanchez, J. Contreras-Garcia, A. J. Cohen and W. Yang, J. Am. Chem. Soc., 2010, 132, 6498–6506 CrossRef CAS PubMed.
- T. Lu and F. Chen, J. Comput. Chem., 2012, 33, 580–592 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 (Revision B.01), Gaussian, Inc., Wallingford CT, 2010 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra05710d |
| This journal is © The Royal Society of Chemistry 2014 |