Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The predominant species of ionic silver in biological media is colloidally dispersed nanoparticulate silver chloride
K. Lozaa,
C. Sengstockb,
S. Chernousovaa,
M. Köllerb and
M. Epple*a
aInorganic Chemistry and Center for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, 45117 Essen, Germany. E-mail: matthias.epple@uni-due.de
bBergmannsheil University Hospital/Surgical Research, Ruhr-University of Bochum, Buerkle-de-la-Camp-Platz 1, 44789 Bochum, Germany
First published on 4th August 2014
Abstract
We have investigated the behaviour of silver ions in biologically relevant concentrations (10 to 100 ppm) in different media, from physiological salt solution over phosphate-buffered saline solution to protein-containing cell culture media. The results show that the initially present silver ions are bound as silver chloride due to the presence of chloride. Only in the absence of chloride, glucose is able to reduce Ag+ to Ag0. The precipitation of silver phosphate was not observed in any case. We conclude that the predominant silver species in biological media is dispersed nanoscopic silver chloride, surrounded by a protein corona which prevents the growth of the crystals and leads to colloidal stabilization. Therefore, in cell culture experiments where dissolved silver ions are studied in the upper ppm range, in fact the effect of colloidally dispersed silver chloride is observed. We have confirmed this by cell culture experiments (human mesenchymal stem cells; T-cells; monocytes) and bacteria (S. aureus) where the cells were incubated with synthetically prepared silver chloride nanoparticles (diameter ca. 100 nm). These were easily taken up by eukaryotic cells and showed the same toxic effect at the same silver concentration as ionic silver (as silver acetate). Therefore, nanoscopic silver chloride and not free ionic silver is the primary toxic species in biological media.
Introduction
Silver as antibacterial agent is increasingly used in many applications.1–4 To meet the diversity of application types, different kinds of silver compound have been developed to serve this market. The most potent compounds for rapid silver release are water-soluble silver salts like silver nitrate or silver acetate. They are often used in cell culture experiments to elucidate the biological effects of silver. In these cases is tacitly assumed that the concentration of free silver ions is the same as that in the added silver salts. This obviously cannot be true because of the presence of a whole set of proteins, biomolecules and inorganic ions like chloride and phosphate in the biological medium. These will react with the silver ions in one or the other way, reducing the concentration of dissolved silver ions, as pointed out by a number of authors,5–7 and also for engineered silver nanoparticles after their release into the environment.8–11Consequently, there have been attempts to study the biological effect of silver chloride5–7,12 which is the most likely precipitation product in the absence of sulphide.2,13 However, in all these studies, silver chloride was prepared as precipitated solid, i.e. not in the presence of proteins and therefore not colloidally stable. Here we show that silver chloride is present as dispersed colloidal species if proteins are present (due to the formation of a stabilizing protein corona)14–20 and that it exhibits the same toxicity towards eukaryotic cells and bacteria as a dissolved silver salt. We deduce that this is the predominant species in “solutions” of soluble silver salts in biological fluids.
Experimental section
Precipitation experiments of silver compounds in biological media
Silver nitrate was added to different biological media with silver ion concentrations between 0.01 g L−1 and 0.1 g L−1 (10 to 100 ppm). This is the range of the cytotoxicity of silver.2,21 The solutions of silver nitrate were stirred at room temperature for 7 days under sterile conditions. Light was not explicitly excluded. All precipitates were isolated by ultracentrifugation (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900g; 30 min), redispersed in pure water, again subjected to ultracentrifugation and then analysed by X-ray powder diffraction, scanning electron microscopy and energy-dispersive X-ray spectroscopy.
900g; 30 min), redispersed in pure water, again subjected to ultracentrifugation and then analysed by X-ray powder diffraction, scanning electron microscopy and energy-dispersive X-ray spectroscopy.
Synthesis of colloidally dispersed silver chloride nanoparticles
Poly(N-vinylpyrrolidone) (PVP)-coated silver chloride nanoparticles were synthesized by precipitation from aqueous solutions of sodium chloride and silver nitrate in the presence of PVP.22 45 mg sodium chloride and 54 mg PVP were dissolved in 12 mL water and heated to 60 °C for 30 min. Then 57 mg AgNO3 dissolved in 1 mL water was quickly added. The dispersion was stirred at 60 °C for 30 min. The particles were collected by ultracentrifugation (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g; 15 min), redispersed in pure water and collected again by ultracentrifugation. The silver chloride nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
400g; 15 min), redispersed in pure water and collected again by ultracentrifugation. The silver chloride nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
Synthesis of fluorescent colloidally dispersed silver chloride nanoparticles
Poly(ethyleneimine) (PEI)-coated silver chloride nanoparticles were synthesized by precipitation from aqueous solutions of sodium chloride and silver nitrate in the presence of PEI by simultaneously pumping aqueous solutions of sodium chloride (30 mg mL−1) and silver nitrate (25 mg mL−1) in a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL into a stirred glass vessel containing 5 mL of PEI solution (2 mg mL−1) during one minute and then stirring for 10 min. Then 50 μL of a solution of fluorescein in ethanol (20 mg mL−1) was added. The dispersion was stirred at room temperature for 120 min. The particles were collected by ultracentrifugation (29
1 mL into a stirred glass vessel containing 5 mL of PEI solution (2 mg mL−1) during one minute and then stirring for 10 min. Then 50 μL of a solution of fluorescein in ethanol (20 mg mL−1) was added. The dispersion was stirred at room temperature for 120 min. The particles were collected by ultracentrifugation (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g; 15 min), redispersed in pure water and collected again by ultracentrifugation. The silver chloride nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
400g; 15 min), redispersed in pure water and collected again by ultracentrifugation. The silver chloride nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
Reagents and methods
Poly(N-vinylpyrrolidone) (PVP K30, Aldrich, molecular weight 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), silver nitrate (Roth, p.a.), sodium chloride (VWR, p.a.), D-(+)-glucose (Fluka, p.a.), fluorescein (Caelo, p.a.) and polyethylenimine (PEI; branched, Aldrich, molecular weight 25
000 g mol−1), silver nitrate (Roth, p.a.), sodium chloride (VWR, p.a.), D-(+)-glucose (Fluka, p.a.), fluorescein (Caelo, p.a.) and polyethylenimine (PEI; branched, Aldrich, molecular weight 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) were used as obtained from the manufacturer. Phosphate-buffered saline solution (PBS) was obtained from Gibco. RPMI 1640 was obtained from Gibco. LB medium was obtained from Sigma-Aldrich. Fetal calf serum (FCS) was obtained from Gibco. Ultrapure water was prepared with an ELGA Purelab ultra instrument.
000 g mol−1) were used as obtained from the manufacturer. Phosphate-buffered saline solution (PBS) was obtained from Gibco. RPMI 1640 was obtained from Gibco. LB medium was obtained from Sigma-Aldrich. Fetal calf serum (FCS) was obtained from Gibco. Ultrapure water was prepared with an ELGA Purelab ultra instrument.
Scanning electron microscopy (SEM) was performed with a FEI Quanta 400 ESEM instrument in high vacuum after sputtering with Au![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd (80
Pd (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20). Energy-dispersive X-ray spectroscopy was carried out with a Genesis 4000 instrument. The hydrodynamic diameter and the zeta potential of the dispersed nanoparticles were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS instrument. The polydispersity index (PDI) was below 0.3 in all cases, indicating a good degree of dispersion. The concentration of silver was determined by atomic absorption spectroscopy (AAS; Thermo Electron Corporation, M-Series). The detection limit was 1 μg L−1 (1 ppb). X-ray powder diffraction was carried out on a Bruker D8 Advance instrument in Bragg–Brentano mode with Cu Kα radiation (1.54 Å). Fluorescence spectroscopy was carried out with a Carry Eclipse fluorescence spectrophotometer. Nanoparticle tracking analysis was performed with a NanoSight LM10 HS instrument.
20). Energy-dispersive X-ray spectroscopy was carried out with a Genesis 4000 instrument. The hydrodynamic diameter and the zeta potential of the dispersed nanoparticles were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS instrument. The polydispersity index (PDI) was below 0.3 in all cases, indicating a good degree of dispersion. The concentration of silver was determined by atomic absorption spectroscopy (AAS; Thermo Electron Corporation, M-Series). The detection limit was 1 μg L−1 (1 ppb). X-ray powder diffraction was carried out on a Bruker D8 Advance instrument in Bragg–Brentano mode with Cu Kα radiation (1.54 Å). Fluorescence spectroscopy was carried out with a Carry Eclipse fluorescence spectrophotometer. Nanoparticle tracking analysis was performed with a NanoSight LM10 HS instrument.
The simulation of the equilibrium of silver species in the biological media was performed with the program Visual MINTEQ 3.0 (J. P. Gustafsson, Stockholm, 2011). The simulation was performed with 0.1 g L−1 (100 ppm) dissolved silver. The media model considered only inorganic salts, no organic compounds. Solids were allowed to precipitate in the model.
Cell culture experiments
Human mesenchymal stem cells (hMSCs, 3rd to 7th passage, Lonza, Walkersville Inc., MD, USA) were cultured in cell culture medium RPMI/10% FCS using 24-well cell culture plates (Falcon, Becton Dickinson GmbH, Heidelberg, Germany). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. hMSCs were sub-cultivated every 7–14 days, depending on the cell proliferation rate. Adherent cells were washed with phosphate-buffered saline solution (PBS, GIBCO, Life Technologies) and detached from the culture flasks by addition of 0.2 mL cm−2 0.25% trypsin/0.1% ethylenediamine tetraacetic acid (EDTA, Sigma-Aldrich, Taufkirchen, Germany) for 5 min at 37 °C. Subsequently, the hMSCs were collected and washed twice with RPMI/10% FCS. Subconfluently growing hMSCs were incubated for 24 h at 37 °C in the presence or absence of different concentrations of AgCl nanoparticles or silver ions (aqueous silver acetate solution), normalized to the silver content, in a humidified atmosphere of 5% CO2. The viability and the morphology of the incubated hMSCs were analyzed using calcein–acetoxymethylester fluorescence staining (calcein–AM, Calbiochem, Schwalbach, Germany). After incubation for 24 h, the AgCl-treated and the silver ion-treated cells were washed twice with RPMI and incubated with calcein–AM (1 μM) at 37 °C for 30 min under cell culture conditions. Subsequently, the adherent cells were washed again with RPMI and analyzed by fluorescence microscopy (Olympus MVX10, Olympus, Hamburg, Germany). Fluorescence microphotographs were taken (Cell P, Olympus) and digitally processed using Adobe Photoshop® 7.0. In this method, living cells give rise to a green fluorescence.Peripheral blood mononuclear cells (PBMC) were isolated by a single-step procedure that was based on a discontinuous double-Ficoll gradient described by English and Andersen.23 Briefly, EDTA-anticoagulated peripheral blood (9 mL Monovette®, Sarstedt, Nürnbrecht, Germany), obtained from healthy volunteers (covered by the approval of the local ethics committee #3036-07), were diluted with an equal volume of 0.9% aqueous NaCl and carefully overlaid on a double gradient formed by layering 10 mL of aqueous polysucrose/sodium diatrizoate, adjusted to a density of 1.077 g mL−1 (Histopaque 1077, Sigma-Aldrich, Taufkirchen, Germany), on 10 mL Histopaque 1119 (Sigma-Aldrich) in 50 mL Falcon tubes (BD-Biosciences, Heidelberg, Germany). The tubes were centrifuged at 700g for 30 min at room temperature. After centrifugation, two distinct leukocyte cell layers (PBMC and polymorphonuclear neutrophil granulocytes, PMN) were obtained above the bottom sediment of erythrocytes. The PBMC layer was carefully aspirated and transferred to a separate 50 mL tube, and the tube was filled with phosphate buffered saline (PBS, Sigma-Aldrich) and centrifuged at 200g for 15 min at 4 °C. This method led to more than 95% pure and viable PBMC. Cell counting was performed using Tuerk staining solution (Sigma-Aldrich). The isolated cells were adjusted to 1 × 106 cells per mL in RPMI 1640 (GIBCO, Invitrogen GmbH, Karlsruhe, Germany) cell culture medium supplemented with L-glutamine (0.3 g L−1), sodium bicarbonate (2.0 g L−1), 10% fetal calf serum (FCS, GIBCO, Invitrogen GmbH), and 20 mM N-(2-hydroxyethyl)-piperazine-N′-(2-ethanesulfonic acid) (HEPES, Sigma-Aldrich). For the differentiation of monocytes and T-cells in the PBMC-fraction, the cells were centrifuged at 370g for 5 min at room temperature. The supernatants were discarded, and the pellets were carefully resuspended. Fluorochrome-labelled anti-CD3 and anti-CD14 (BD Bioscience, Heidelberg, Germany) were added (20 μL each), and the samples were incubated for 30 min in the dark at room temperature. Subsequently, the cells were washed with PBS, centrifuged at 370g for 5 min at RT and fixed with 1.5% formaldehyde.
HeLa cells were cultivated in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal calf serum (FCS) at 37 °C in humidified atmosphere with 5% CO2. Approximately 12 h prior to the addition of the silver compounds, the cells were seeded into a six-well plate over cover glass. The silver concentration in cell culture medium was 2 μg mL−1 (2 ppm). The cells were incubated for 4 h under cell culture conditions, then washed three times with PBS, and studied with a fluorescence microscope BZ-9000 (Keyence, Japan).
Bacterial culture experiments
Bacterial tests were performed with Staphylococcus aureus (DSMZ 1104), obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures). S. aureus was grown in BHI broth (brain–heart infusion broth, bioMérieux, Nürtingen, Germany) overnight at 37 °C in a water bath. Bacterial concentrations of overnight cultures were measured using a Densichek® (bioMérieux, Lyon, France) turbidity photometer. The calculation of bacterial counts was based on turbidity standard solutions (McFarland scale).The antimicrobial activity of nanoparticulate silver chloride was tested using standard methods which determine the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC). MIC was determined in RPMI 1640 (Life Technologies) containing 10% (v/v) fetal calf serum (FCS, Life Technologies) and L-glutamine (0.3 g L−1, Life Technologies), and defined as the lowest silver concentration able to inhibit bacterial growth (no visible growth) in 2 mL plastic macrodilution test tubes. Therefore, working silver stock solutions (50 μL) were added to 1 mL of the respective liquid culture medium, and different cell numbers (103 to 106 mL−1) of bacteria were used for inoculation. Cells were incubated in a cell culture incubator (RPMI/10% FCS) in the presence of 5% carbon dioxide in a humidified atmosphere at 37 °C overnight.
The minimum bactericidal concentration (MBC) was subsequently determined by plating 100 μL aliquots of the MIC samples on blood agar plates. The MBC was defined as the lowest silver concentration which completely prevented colony forming units (CFU) on the agar plate.
Statistical analysis
Data are expressed as mean ± standard deviation of at least three independent experiments. Analysis of the data distribution was performed using Student's t-test to analyse the significance of differences between the treated group and the control group (without silver exposure). P values of less than 0.05 were considered to be statistically significant.Results and discussion
Biological media always contain considerable amounts of chloride which leads to the precipitation of silver chloride. The equilibrium concentration of free ionic silver can be calculated using the solubility product of AgCl (1.7 × 10−10 mol2 L−2).2 However, ionic silver can form complexes with organic compounds and thereby be removed from the equilibrium. This leads to an increased solubility. Furthermore, precipitating silver chloride may be rapidly coated by biomolecules and prevented from further growth due to the forming protein corona.15,16,24–26 However, the concentration of ionic silver is typically low in biological experiments (between 1 and 100 ppm). This may explain why a precipitation is typically not observed: the resulting particles are very small and do not precipitate, and their overall concentration is low. In the following, we will elaborate the argumentation chain about the state of silver ions in biological media.First, we have added silver nitrate in a low concentration (10 to 100 ppm) to various biological media. Such a concentration is in the range of biological effects of silver ions.2 The dispersions were stirred for 7 days, the resulting solid products were isolated by ultracentrifugation and then identified by X-ray diffraction, scanning electron microscopy and energy-dispersive X-ray spectroscopy. Note that a gravimetric determination of the amount of precipitate was impossible due to the very small amount (about 100–150 mg from 1 L of dispersion). This also prevented the analysis of even more lower silver concentrations. In all cases, we found sub-μm-sized particles of silver chloride (Table 1). The only exception was the aqueous solution of glucose, i.e. a reducing agent, where we found metallic silver. Notably, the particles were smaller if proteins were present, indicating an inhibition of the crystal growth by a protein corona. This is reasonable because the culture media contain about 15 g L−1 (LB) and 10 g L−1 (RPMI/10% FCS) of protein, respectively. The analysis of the nature of the protein corona is difficult and beyond the scope of this manuscript. In the absence of biomolecules, the silver chloride crystals had the possibility to grow until the supersaturation had been consumed by crystallization. No metallic silver was found by X-ray diffraction together with silver chloride, indicating that any reduction (e.g. by light) was either minor or absent. Test experiments with a shorter immersion time (12 h) gave identical results, i.e. the precipitation and the equilibration of AgCl occur within a few hours (data not shown).
| Medium (aqueous solutions) | Diameter of the particles by SEM | Maximum possible concentration of Ag+ in the immersion medium/μg L−1 | Concentration of Cl−/M |
|---|---|---|---|
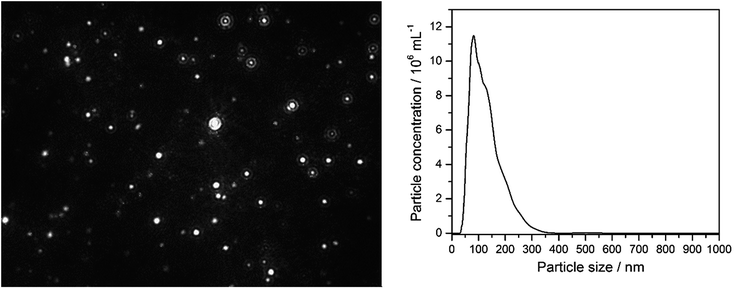
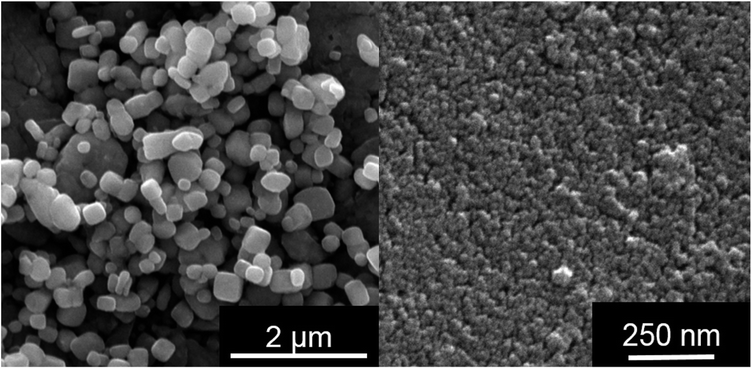
| a The dispersed nanoparticles particles were also directly detected by nanoparticle tracking analysis (NTA) in RPMI/10% FCS (Fig. 1). This was not possible in LB medium due to the high turbidity and the small silver chloride crystals. Fig. 2 shows SEM images of AgCl nanoparticles, isolated from PBS (i.e. no protein present) and LB medium, respectively. | |||
| Glucose (2 g L−1) | 50–100 nm (Ag0) | — | 0 |
| NaCl (0.9%) | 300–1000 nm (AgCl) | 0.225 | 0.154 |
| NaCl (0.9%) + glucose (2 g L−1) | 700–1500 nm (AgCl) | 0.225 | 0.154 |
| PBS | 500–1000 nm (AgCl) | 0.250 | 0.139 |
| PBS + glucose (2 g L−1) | 800–1500 nm (AgCl) | 0.250 | 0.139 |
| RPMI 1640 medium/10% FCS | 200–350 nm (AgCl) | 0.327 | 0.108 |
| LB medium | 30–50 nm (AgCl) | 0.372 | 0.085 |
Having shown that nanoscopic particles of AgCl form in biological media, we wanted to test the biological activity of such AgCl nanoparticles. Silver chloride nanoparticles were synthetically prepared and stabilized by polyelectrolytes (PVP or PEI, respectively). Table 2 lists the colloid-chemical data of the prepared silver chloride nanoparticles in comparison to those formed in the biological media. Fig. 3 gives dynamic light scattering data of PVP-stabilized AgCl nanoparticles.
| Size by DLS/nm | Size by SEM/nm | Zeta-potential/mV | Size by NTA/nm | |
|---|---|---|---|---|
| AgCl in LB | — | 30–50 | — | — |
| AgCl in RPMI + 10% FCS | — | 200–350 | — | 50–350 |
| AgCl–PVP | 120 | 100 | −42 | 110 |
| AgCl–PEI | 220 | 80 | +67 | 180 |
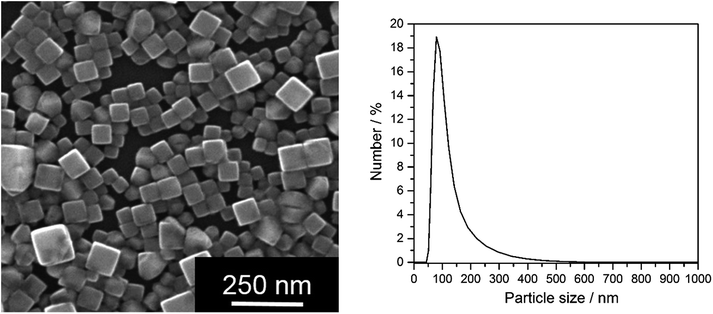 | ||
| Fig. 3 Synthetically prepared AgCl–PVP nanoparticles. Left: SEM image; right: particle size distribution (by number) from DLS of water-dispersed particles. | ||
The synthetically prepared AgCl–PVP nanoparticles were also colloidally stable in RPMI + 10% FCS for at least one week (Fig. 4).
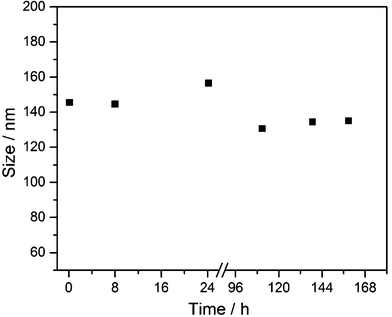 | ||
| Fig. 4 Colloidal stability of synthetic AgCl–PVP nanoparticles, dispersed in RPMI + 10% FCS medium at 37 °C under sterile conditions, by dynamic light scattering (DLS). No precipitation was observed. | ||
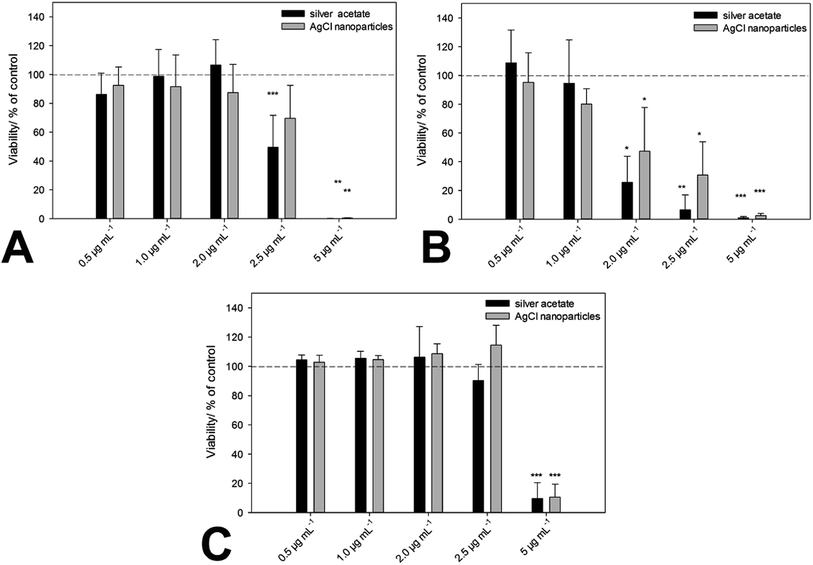
Up to this point, we have identified nanoscopic AgCl nanoparticles in biological media, and also prepared comparable particles which are colloidally stable in biological media. The next step is the comparison of the biological effect of “naturally occurring” AgCl nanoparticles and “synthetic” AgCl nanoparticles. This was done with three cell lines (hMSC, T-cells, and monocytes) and a relevant bacterial strain (S. aureus). The cells were incubated with the same amounts of silver, either as soluble silver acetate or as colloidally stable AgCl–PVP nanoparticles. Fig. 5 and Table 3 summarize all results.
| Cell density/mL−1 | Silver acetate (N = 3) | AgCl nanoparticles (N = 3) |
|---|---|---|
| MBC/μg mL−1 Ag | MBC/μg mL−1 Ag | |
| 1 × 103 | 2.0 | 2.0 |
| 1 × 104 | 2.0 | 2.5 to 2.0 |
| 1 × 105 | 2.5 | 5.0 |
| 1 × 106 | 5.0 | 5.0 |
We can see that there is no significant difference between the biological action of silver ions and AgCl-NP towards eukaryotic cells and bacteria. As we have shown earlier, the cytotoxic concentration of silver acetate against eukaryotic cells and bacteria is in the same concentration range, and Gram-negative bacteria are affected by silver ions in the same way as Gram-positive bacteria.21 This underscores the presence and the biological effect of dispersed AgCl nanoparticles in both cases. Finally, we have prepared fluorescing nanoparticles of silver chloride to follow their pathway into cells. HeLa cells were chosen as suitable model cell line. Fig. 6 shows the characterization of the AgCl nanoparticles.
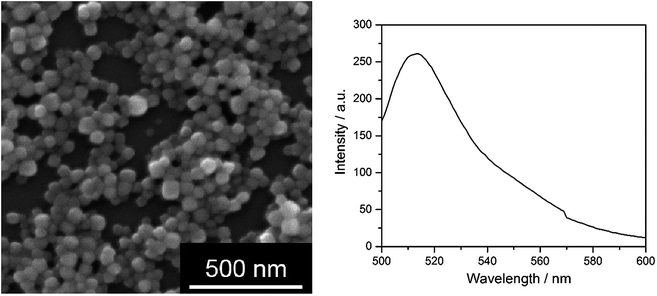 | ||
| Fig. 6 Fluorescing AgCl nanoparticles, stabilized with PEI and labelled with fluorescein. Left: SEM image; right: fluorescence spectrum, excitation wavelength 490 nm. | ||
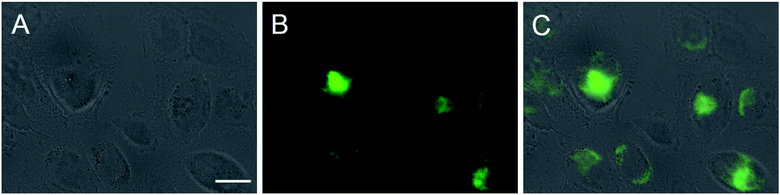
Fig. 7 shows the cellular uptake into HeLa cells which occurred within a few hours. It may be safely assumed that the uptake occurs by endocytosis as in the case of other nanoparticles.27–29
A final question remains: what is the origin of the cytotoxic effect of the AgCl nanoparticles? We clearly have to distinguish between bacteria and eukaryotic cells. The uptake of nanoparticles by bacteria is limited to very small (1–10 nm) particles.30,31 The attachment of these particles to the surface of Gram-negative bacteria may disturb their proper function such as permeability, additionally they may be able to penetrate the cell membrane and cause further damage to a variety of biomolecules e.g. by binding to sulphur-containing molecules or by the release of silver ions.30,31 Thus, AgCl can be effective from the outside, e.g. by disrupting the bacterial cell membrane and also by inducing internal collateral damage like the formation of reactive oxygen species.31 The larger silver chloride nanoparticles that we have found and prepared here are probably acting from inside the cells due to the release of silver ions, as it has been recently shown for silver nanoparticles.32
If AgCl nanoparticles are taken up into eukaryotic cells by endocytosis, they end up in the lysosome under acidic conditions. In principle, the solubility of AgCl should not depend on the pH, with HCl being a strong acid. We have simulated the conditions inside a lysosome, using the software Visual MinteQ 3.0. As lysosomal liquid, we assumed hydrochloric acid with a pH of 5.33 The calculation was performed for 100 μM silver chloride. In this case, about 10% of the AgCl is present in solution as silver ion Ag+, with a small amount of the chloro-complex [AgCl2]− also present. Therefore so far, the question cannot be answered whether AgCl fully dissolves in lysosomes. Except for the formation of the chloro-complex and the unknown degree of complexation by biomolecules, the solubility of AgCl should not be increased inside a lysosome. However, it must be stressed that the solubility product for AgCl was derived for macroscopic crystals. As nanocrystals have a much higher specific surface area, their solubility will generally be higher, and hence their cytotoxicity will be higher than that of AgCl microcrystals.
Conclusions
In chloride-containing media, silver ions are precipitated as silver chloride. If high amounts of proteins are present, as it is typically the case in cell culture media and biological fluids like blood, the silver chloride particles remain in a nanoparticulate state, surrounded and preserved by a protein corona. These particles show the same cytotoxicity as silver ions towards eukaryotic cells and bacteria. They are taken up by eukaryotic cells within a few hours. Although the mechanism of the cytotoxicity of dispersed silver chloride nanoparticles cannot be fully explained so far, the particles are probably acting both from the outside and, if they are taken up by eukaryotic cells, the inside of the cells. The size of formed nanoparticles and aggregation state could also affect the cytotoxicity.5 These considerations should also apply to environmental systems like salt water with high chloride content, provided that biomolecules are present to form a stabilizing corona around the nanoparticles.12,34,35Acknowledgements
We thank the Deutsche Forschungsgemeinschaft (DFG) for financial support of this project within the Priority Program NanoBioResponses (SPP 1313).Notes and references
- J. K. Schluesener and H. J. Schluesener, Arch. Toxicol., 2013, 87, 569–576 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- B. Simoncic and B. Tomsic, Text. Res. J., 2010, 80, 1721–1737 CrossRef CAS PubMed.
- J. W. Alexander, Surg. Infect., 2009, 10, 289–292 CrossRef PubMed.
- S. K. Zhang, C. Du, Z. Z. Wang, X. G. Han, K. Zhang and L. H. Liu, Toxicol. In Vitro, 2013, 27, 739–744 CrossRef CAS PubMed.
- O. Choi, K. K. Deng, N. J. Kim, L. Ross, R. Y. Surampalli and Z. Q. Hu, Water Res., 2008, 42, 3066–3074 CrossRef CAS PubMed.
- H. T. Ratte, Environ. Toxicol. Chem., 1999, 18, 89–108 CrossRef CAS PubMed.
- H. Zhang, J. A. Smith and V. Oyanedel-Craver, Water Res., 2012, 46, 691–699 CrossRef CAS PubMed.
- B. Nowack, J. F. Ranville, S. Diamond, J. A. Gallego-Urrea, C. Metcalfe, J. Rose, N. Horne, A. A. Koelmans and S. J. Klaine, Environ. Toxicol. Chem., 2012, 31, 50–59 CrossRef CAS PubMed.
- J. Y. Liu, Z. Y. Wang, F. D. Liu, A. B. Kane and R. H. Hurt, ACS Nano, 2012, 6, 9887–9899 CrossRef CAS PubMed.
- J. M. Zook, S. E. Long, D. Cleveland, C. L. A. Geronimo and R. I. MacCuspie, Anal. Bioanal. Chem., 2011, 401, 1993–2002 CrossRef CAS PubMed.
- M. Grosell, C. Hogstrand, C. M. Wood and H. J. M. Hansen, Aquat. Toxicol., 2000, 48, 327–342 CrossRef CAS.
- K. Loza, J. Diendorf, C. Greulich, L. Ruiz-Gonzales, J. M. Gonzalez-Calbet, M. Vallet-Regi, M. Koeller and M. Epple, J. Mater. Chem. B, 2014, 2, 1634–1643 RSC.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- V. Mirshafiee, M. Mahmoudi, K. Lou, J. Cheng and M. L. Kraft, Chem. Commun., 2013, 49, 2557–2559 RSC.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- M. P. Monopoli, F. B. Bombelli and K. A. Dawson, Nat. Nanotechnol., 2011, 6, 11–12 CrossRef CAS.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- S. Kittler, C. Greulich, J. S. Gebauer, J. Diendorf, L. Treuel, L. Ruiz, J. M. Gonzalez-Calbet, M. Vallet-Regi, R. Zellner, M. Köller and M. Epple, J. Mater. Chem., 2010, 20, 512–518 RSC.
- C. Greulich, D. Braun, A. Peetsch, J. Diendorf, B. Siebers, M. Epple and M. Koller, RSC Adv., 2012, 2, 6981–6987 RSC.
- C. An, S. Peng and Y. Sun, Adv. Mater., 2010, 22, 2570–2574 CrossRef CAS PubMed.
- D. English and B. R. Andersen, J. Immunol. Methods, 1974, 5, 249–252 CrossRef CAS.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. U. Nienhaus, ACS Nano, 2014, 28, 503–513 CrossRef PubMed.
- J. S. Gebauer, M. Malissek, S. Simon, S. K. Knauer, M. Maskos, R. H. Stauber, W. Peukert and L. Treuel, Langmuir, 2012, 28, 9673–9679 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- T. G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176–185 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182–195 CrossRef CAS PubMed.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramirez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed.
- O. Choi and Z. Q. Hu, Environ. Sci. Technol., 2008, 42, 4583–4588 CrossRef CAS.
- E. Caballero-Díaz, C. Pfeiffer, L. Kastl, P. Rivera-Gil, B. Simonet, M. Valcárcel, J. Jiménez-Lamana, F. Laborda and W. J. Parak, Part. Part. Syst. Charact., 2013, 30, 1079–1085 CrossRef PubMed.
- J. Ji, N. Rosenzweig, C. Griffin and Z. Rosenzweig, Anal. Chem., 2000, 72, 3497–3503 CrossRef CAS.
- J. R. Reinfelder and S. I. Chang, Environ. Sci. Technol., 1999, 33, 1860–1863 CrossRef CAS.
- C. M. Wood, M. Grosell, C. Hogstrand and H. Hansen, Aquat. Toxicol., 2002, 56, 197–213 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |