Inflammatory modulation of stem cells by Magnetic Resonance Imaging (MRI)-detectable nanoparticles†
Sezin Aday‡
ab,
Jose Paiva‡ab,
Susana Sousab,
Renata S. M. Gomesc,
Susana Pedreirod,
Po-Wah Soe,
Carolyn Ann Carrf,
Lowri Cochling,
Ana Catarina Gomesb,
Artur Paivad and
Lino Ferreira*ab
aCNC – Center for Neurosciences and Cell Biology, University of Coimbra, Coimbra, Portugal
bBiocant, Biotechnology Innovation Center, Cantanhede, Portugal. E-mail: lino@biocant.pt
cKing's BHF Centre of Excellence, Cardiovascular Proteomics, King's College London, London, UK
dCentro de Histocompatibilidade do Centro, Coimbra, Portugal
eDepartment of Neuroimaging, Institute of Psychiatry, King's College London, London, UK
fCardiac Metabolism Research Group, Department of Physiology, Anatomy & Genetics, University of Oxford, UK
gPulseTeq Limited, Chobham, Surrey, UK
First published on 2nd July 2014
Abstract
In the current work, we labelled human hematopoietic stem cells with polymeric nanoparticles (NPs) that can be tracked by Magnetic Resonance Imaging (MRI) and studied their effect on cell metabolism, proliferation, secretomics, genomics and differentiation. We showed that NPs had no effect on the stem cell differentiation program but affected their paracrine activity.
Hematopoietic stem cells (HSCs) are being evaluated in several clinical trials for the treatment of the heart after angina1 or infarction,2 chronic wounds,3,4 ischemic limb,5,6 etc.… MRI is the most attractive imaging modality to track cells because it provides high-quality 3-dimensional functional and anatomic information with high contrast.7 Emulsions containing fluorine are being used to label different type of cells since there is no fluorine in the human body, and therefore cells labeled with these NPs can be selectively imaged by 19F MRI.8–10 Unfortunately, so far it is unclear the effect of the NPs in the biology of stem cells, particularly in their differentiation program as well as their paracrine activity. Studies have shown that superparamagnetic iron NPs coated with protamine sulfate had no measurable cytotoxicity on HSCs but their effect in the differentiation and paracrine activity of HSCs was not demonstrated.11 In addition, self-assembling ferumoxytol–heparin protamine nanocomplexes were also used as labeling agents for MRI.12 The internalization of these nanocomplexes by HSCs (1.33 ± 0.2 pg iron per cell) slightly decreased the proliferation of HSCs and proliferative capacity of the cells was recovered only after 30 days.
Here we studied the effect of poly(lactic acid-co-glycolic acid) (PLGA) NPs containing perfluoro-1,5-crown ether (PFCE) with an average diameter of 210 nm (therefore named as NP210-PFCE) and polydispersity index of 0.12 ± 0.01 (Fig. 1A), that have the capacity to track cells,13 on the viability, proliferation, secretomics, genomics and differentiation of hematopoietic stem/progenitor cells (CD34+ cells). Endothelial cells (ECs) have been used as control. Initially, internalization studies were performed using fluorescence activated cell sorting (FACS) and fluorimetry to calculate the percentage of the cells labeled and the amount of NPs internalized by the cells, respectively. Cytotoxicity profile of the NP210-PFCE formulation was evaluated by cell counting, ATP production, LDH release, ROS production, secretion of pro-inflammatory cytokines and gene expression. Then, the effect of NP formulation on the differentiation capacity of CD34+ HSCs was evaluated. Finally, the mechanism underlying the anti-inflammatory effect of our NPs was studied. Our results show that NPs have no substantial effect in cell proliferation, metabolism and differentiation of CD34+ cells or ECs; however, the NPs significantly reduced the inflammatory activity of CD34+ cells but not ECs, as evaluated by gene microarray and protein secretion analysis.
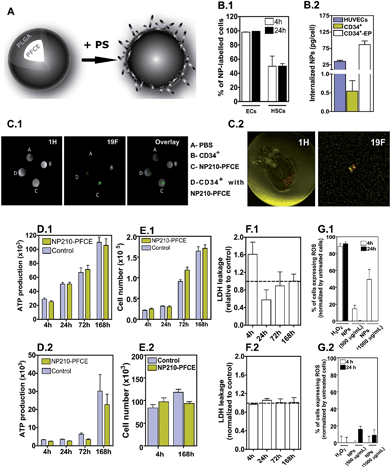
NP210-PFCE containing 176.5 μg of PFCE per mg of PLGA was coated with protamine sulfate (PS; approximately 13 μg of PS per mg of NP), a cationic agent to facilitate intracellular delivery (Fig. 1A).11 In these conditions, NP210-PFCE had an average diameter of 210 nm and a zeta potential of 7.0 ± 1.7 mV. HUVECs and CD34+ cells were exposed to fluorescent-labeled NPs (500 μg mL−1) for 4 h, harvested and labeling efficiency of the cells was assessed by Fluorescence Activated Cell Sorting (FACS) and fluorimetry. FACS results show that 95% (ECs) and 50% (CD34+) of the cells were labeled after 4 h (Fig. 1B.1). Fluorescence measurements indicate that this labeling corresponds to the internalization of 34.7 ± 3.4 and 0.5 ± 0.3 pg of PFCE per cell of HUVECs and CD34+, respectively (Fig. 1B.2). When the CD34+ cells were electroporated and incubated with higher concentrations (2 mg mL−1) of NPs up to 24 h, the internalized amount of NPs increased up to 86.4 ± 11.2 pg of PFCE per cell in vitro (Fig. 1B.2). Under this loading condition, which is comparable to fluorine-based liposomes used in the literature for different cell types,8 the cells can be tracked by MRI (Fig. 1C.1) However, MRI can also monitor CD34+ cells in vivo without electroporation (Fig. 1C.2).
NP210-PFCE internalized by CD34+ cells or ECs have no substantial effect in cell proliferation, metabolism and oxidative stress. Cells labeled with fluorescent NPs were sorted by flow cytometry before evaluation. Both cells transfected with NP210-PFCE show similar cell metabolism, as assessed by an ATP assay, and cell proliferation as compared to untreated cells (Fig. 1D and E). In addition, after 24 h, no significant differences were observed in LDH release on CD34+ cells or HUVECs treated with or without NPs, showing that NPs do not significantly affect cell membrane integrity (Fig. 1F). Moreover, CD34+ cells cultured with NPs show low capacity to generate ROS even after 24 h of culture (Fig. 1G). The relative low effect of NPs in ROS generation is likely due to CD34+ cell capacity to tolerate oxidative stress due to high expression and activity of antioxidant enzymes (see below).14 This phenomenon might explain the toleration of the cells to NPs and H2O2. ECs cultured for 4 h in the presence of NP210-PFCE generate ROS but then the levels drop off to baseline levels if cells were cultured for additional 20 h in the absence of NPs. Together, our results indicate that the oxidative stress induced by NPs was moderate in CD34+ cells and ECs and the increase in ROS was time-, dose- and cell-dependent.
NP210-PFCE formulation has no effect on the endothelial differentiation capacity of CD34+ cells. CD34+ cells transfected with NP210-PFCE for 4 h were then sorted to isolate the ones containing NPs. Both untreated and NP-treated CD34+ cells attached to the culture dish after 15–20 days of culture with a cobblestone-like morphology and expressed high levels of EC markers4 (Fig. 2). No differences were observed in terms of kinetics or phenotype between both cells. Overall, our results indicate that NP210-PFCE has no measurable effect in the in vitro differentiation program of CD34+ cells.
Next, we investigated the impact of the NPs on both cells by gene microarray analysis. Gene expression in HUVECs was not statistically significant in cells treated or not with NPs. In contrast, CD34+ cells showed more than 100 genes that were statistically (p < 0.001) significant and grouped in 5 categories and 15 sub-categories (Fig. 3A). Interestingly, the most affected cellular function was immune response, being 41 genes downregulated at 24 h, and 7 genes at day 7 (ESI Tables 2–8†). In addition, there was an increase in the expression of the genes that prevent oxidative stress, in particular metallothioneins (MTs).15
Some of the genes identified in the microarray analysis have been confirmed by qRT-PCR (Fig. 3B). The cellular expression of MTs at mRNA level is dependent on the initial concentration of the NPs that cells were exposed to (Fig. 3C). Importantly, the anti-inflammatory properties of NP210-PFCE are not shared by superparamagnetic iron oxide NPs (SPION), very often used in MRI applications, which have a similar internalization level (0.27 ± 0.01 pg per cell) as NP210-PFCE (Fig. 3B). To confirm the impact of the NPs, we evaluated the secretion of pro-inflammatory cytokines in cells exposed to NPs for 24 h by Bioplex. CD34+ cells incubated with NPs show a decrease in the secretion of pro-inflammatory cytokines (e.g. IFN-γ, IL-8, MCP-1, MIP-1β, TNF-α) (Fig. 3D). In contrast, HUVECs containing NPs increased the secretion of IL-6 (∼2.0 fold) and MCP-1 (∼1.5 fold) among all cytokines tested (Fig. 3E). Altogether, our results show that the exquisite properties (chemistry (PLGA, PS or PFCE), size or geometry) of NP210-PFCE are unique in inducing the anti-inflammatory properties on CD34+ cells. Further testing is needed to elucidate this issue. Although not shown, soluble protamine sulfate has no anti-inflammatory properties on CD34+ cells.
NP210-PFCE formulation interferes with agonist-mediated activation of TLRs. TLRs are pattern-recognition receptors that allow cells to recognize and protect tissues from various harmful stimuli.16 At least 10 human TLRs have been identified so far, being TLRs 1, 2, 4, 5 and 6 mainly located on the cell surface while TLRs 3, 7, 8 and 9 mostly found in the endocytic compartments.17 Cellular activation of TLRs leads to the expression of inflammatory cytokines and chemokines. CD34+ cells express 9 TLRs at mRNA level, being TLR2 and TLR-7 the lowest expressed (Fig. 3F). Next, we evaluated whether CD34+ cells labeled with NP210-PFCE showed attenuated activation of agonist-mediated activation of TLRs. From all the TLRs tested, the activation of TLR6 and TLR7 by TLR agonists was significantly decreased (p < 0.001 for TLR6 and p < 0.05 for TLR7) (Fig. 3G). Overall, our results indicate that the immunomodulatory properties of the NP210-PFCE are mediated by TLR6 and TLR7.
Conclusions
This work shows that NPs with the capacity to track stem cells may have immunomodulatory properties. Previous studies have used several NP formulations to track CD34+ cells in vivo (e.g. superparamagnetic18 as well as nanocomplexes12) by MRI; however, the effects of NPs in stem cell biology were never addressed using high-throughput characterization techniques such as gene arrays. Our results show that NPs may interfere with CD34+ TLRs attenuating their secretion of inflammatory cytokines. Although studies have shown that NPs (specifically silver NPs) can inhibit the secretion of pro-inflammatory cytokines (IL-6, IL-8, TNF-α, etc.…) on macrophages mediated by specific TLRs,19 such studies were never extended to HSCs and more specifically to NPs with the capacity to track stem cells. The immunomodulatory properties of NPs may be relevant in a biomedical context since it has been shown that inhibition of TLR and proinflammatory cytokine signaling contributes critically to ischemic tolerance in different organs.20,21 Injured tissue and necrotic cells release endogenous activators (TLR ligands),22 which activate TLRs and causes of high level of inflammatory cytokine secretion and cellular injury. However, down-regulation of proinflammatory TLR and cytokine signaling reduces the acute inflammatory response that worsens ischemic injury. Different studies showed that both TLR2- and TLR4-deficient mice show less injury upon cerebral ischemia.23 Similarly, our NPs might be an interesting tool to increase ischemic tolerance and cell survival in different organs. Altogether, the NPs described in this work represent a novel class of NPs for MRI imaging while acting as an anti-inflammatory agent.Acknowledgements
This work was supported by a Marie Curie-Reintegration Grant (FP7-People-2007-4-3-IRG; contract no. 230929), MIT-Portugal program, FCT (PTDC/CTM/099659/2008; and SFRH/BD/42871/2008, a fellowship to S.A.) and COMPETE funding (Project “Stem cell based platforms for Regenerative and Therapeutic Medicine”, Centro-07-ST24-FEDER-002008).Notes and references
- D. W. Losordo, R. A. Schatz, C. J. White, J. E. Udelson, V. Veereshwarayya, M. Durgin, K. K. Poh, R. Weinstein, M. Kearney, M. Chaudhry, A. Burg, L. Eaton, L. Heyd, T. Thorne, L. Shturman, P. Hoffmeister, K. Story, V. Zak, D. Dowling, J. H. Traverse, R. E. Olson, J. Flanagan, D. Sodano, T. Murayama, A. Kawamoto, K. F. Kusano, J. Wollins, F. Welt, P. Shah, P. Soukas, T. Asahara and T. D. Henry, Circulation, 2007, 115, 3165–3172 CrossRef PubMed.
- V. Schachinger, S. Erbs, A. Elsasser, W. Haberbosch, R. Hambrecht, H. Holschermann, J. Yu, R. Corti, D. G. Mathey, C. W. Hamm, T. Suselbeck, B. Assmus, T. Tonn, S. Dimmeler and A. M. Zeiher, N. Engl. J. Med., 2006, 355, 1210–1221 CrossRef CAS PubMed.
- O. Awad, E. I. Dedkov, C. Jiao, S. Bloomer, R. J. Tomanek and G. C. Schatteman, Arterioscler., Thromb., Vasc. Biol., 2006, 26, 758–764 CrossRef CAS PubMed.
- D. C. Pedroso, A. Tellechea, L. Moura, I. Fidalgo-Carvalho, J. Duarte, E. Carvalho and L. Ferreira, PLoS One, 2011, 6, e16114 CAS.
- R. J. Powell, W. A. Marston, S. A. Berceli, R. Guzman, T. D. Henry, A. T. Longcore, T. P. Stern, S. Watling and R. L. Bartel, Mol. Ther., 2012, 20, 1280–1286 CrossRef CAS PubMed.
- R. Passier, L. W. van Laake and C. L. Mummery, Nature, 2008, 453, 322–329 CrossRef CAS PubMed.
- A. Stroh, C. Faber, T. Neuberger, P. Lorenz, K. Sieland, P. M. Jakob, A. Webb, H. Pilgrimm, R. Schober, E. E. Pohl and C. Zimmer, NeuroImage, 2005, 24, 635–645 CrossRef PubMed.
- E. T. Ahrens, R. Flores, H. Xu and P. A. Morel, Nat. Biotechnol., 2005, 23, 983–987 CrossRef CAS PubMed.
- M. Srinivas, P. A. Morel, L. A. Ernst, D. H. Laidlaw and E. T. Ahrens, Magn. Reson. Med., 2007, 58, 725–734 CrossRef CAS PubMed.
- Y. B. Yu, J. Drug Targeting, 2006, 14, 663–669 CrossRef CAS PubMed.
- A. S. Arbab, G. T. Yocum, H. Kalish, E. K. Jordan, S. A. Anderson, A. Y. Khakoo, E. J. Read and J. A. Frank, Blood, 2004, 104, 1217–1223 CrossRef CAS PubMed.
- M. S. Thu, L. H. Bryant, T. Coppola, E. K. Jordan, M. D. Budde, B. K. Lewis, A. Chaudhry, J. Ren, N. R. Varma, A. S. Arbab and J. A. Frank, Nat. Med., 2012, 18, 463–467 CrossRef CAS PubMed.
- R. S. Gomes, R. P. Neves, L. Cochlin, A. Lima, R. Carvalho, P. Korpisalo, G. Dragneva, M. Turunen, T. Liimatainen, K. Clarke, S. Yla-Herttuala, C. Carr and L. Ferreira, ACS Nano, 2013, 7, 3362–3372 CrossRef CAS PubMed.
- T. He, T. E. Peterson, E. L. Holmuhamedov, A. Terzic, N. M. Caplice, L. W. Oberley and Z. S. Katusic, Arterioscler., Thromb., Vasc. Biol., 2004, 24, 2021–2027 CrossRef CAS PubMed.
- S. K. Baird, T. Kurz and U. T. Brunk, Biochem. J., 2006, 394, 275–283 CrossRef CAS PubMed.
- J. Lee, J. W. Sohn, Y. Zhang, K. W. Leong, D. Pisetsky and B. A. Sullenger, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 14055–14060 CrossRef CAS PubMed.
- S. Akira, S. Uematsu and O. Takeuchi, Cell, 2006, 124, 783–801 CrossRef CAS PubMed.
- M. Lewin, N. Carlesso, C. H. Tung, X. W. Tang, D. Cory, D. T. Scadden and R. Weissleder, Nat. Biotechnol., 2000, 18, 410–414 CrossRef CAS PubMed.
- P. M. Castillo, J. L. Herrera, R. Fernandez-Montesinos, C. Caro, A. P. Zaderenko, J. A. Mejias and D. Pozo, Nanomedicine, 2008, 3, 627–635 CrossRef PubMed.
- K. Kariko, D. Weissman and F. A. Welsh, J. Cereb. Blood Flow Metab., 2004, 24, 1288–1304 CrossRef CAS PubMed.
- Y. C. Wang, S. Lin and Q. W. Yang, J. Neuroinflammation, 2011, 8, 134 CrossRef CAS PubMed.
- Y. Shi and K. L. Rock, Eur. J. Immunol., 2002, 32, 155–162 CrossRef CAS.
- S. C. Tang, T. V. Arumugam, X. Xu, A. Cheng, M. R. Mughal, D. G. Jo, J. D. Lathia, D. A. Siler, S. Chigurupati, X. Ouyang, T. Magnus, S. Camandola and M. P. Mattson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13798–13803 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Details on experimental methods and supplementary tables. See DOI: 10.1039/c4ra04041d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)