Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family†
Zhi-Biao Wang,
Xi Chen,
Wei Wang,
Ke-Di Cheng and
Jian-Qiang Kong*
Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College (State Key Laboratory of Bioactive Substance and Function of Natural Medicines & Ministry of Health Key Laboratory of Biosynthesis of Natural Products), Beijing, 100050, China. E-mail: jianqiangk@imm.ac.cn; Tel: +86-10-63165169
First published on 22nd May 2014
Abstract
OSW-1 is a promising antitumor glycoside present in the Ornithogalum saundersiae plant. Biosynthesis of the p-methoxybenzoyl group on the disaccharide moiety of OSW-1 is known to take place biochemically by phenylpropanoid biosynthetic pathway, but molecular biological characterization of related genes has been insufficient. Phenylalanine ammonia lyase (PAL, EC 4.3.1.24), which catalyzes the deamination of L-phenylalanine to yield trans-cinnamic acid, plays a key role in phenylpropanoid metabolism. Thus, the study on the characterization of the genes involved in the OSW-1 biosynthetic pathway, particularly the well-documented genes such as PAL, is essential to further the understanding of the biosynthesis of OSW-1. Here, transcriptomic sequencing of O. saundersiae was performed to speed up the identification of a large number of genes related to OSW-1 biosynthesis. De novo assembly of the transcriptome sequence provided 210![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 contigs, 104, 180 unigenes, and four unigenes showing high similarities with PALs. Two full-length cDNAs encoding PALs (OsaPAL2 and OsaPAL62) from O. saundersiae were cloned using sequence information from these four unigenes. The PAL and tyrosine ammonia lyase (TAL) activities of recombinant OsaPAL proteins were unambiguously determined by HPLC with UV and MS detection as well as by NMR spectroscopy. Subsequently, a series of site-directed mutants were generated with the aim of improving enzyme activity and investigating the importance of particular residues in determining substrate selectivity. The results reveal that the Phe-to-His mutants, i.e., OsaPAL2F134H and OsaPAL62F128H, exhibited higher TAL activity than the corresponding wild types, providing direct evidence that the Phe residue is responsible for substrate specificity. Mutagenesis studies also demonstrated that the Thr-to-Ser mutants, i.e., OsaPAL2T196S and OsaPAL62T194S, showed significantly higher substrate affinity than the wild types. Furthermore, the Gly-to-Ala mutants, i.e., OsaPAL2G209A and OsaPAL62G207A, showed higher PAL and TAL activities. These findings provide further insight into the genes responsible for OSW-1 biosynthesis and will facilitate the future application of OsaPALs in synthetic biology.
733 contigs, 104, 180 unigenes, and four unigenes showing high similarities with PALs. Two full-length cDNAs encoding PALs (OsaPAL2 and OsaPAL62) from O. saundersiae were cloned using sequence information from these four unigenes. The PAL and tyrosine ammonia lyase (TAL) activities of recombinant OsaPAL proteins were unambiguously determined by HPLC with UV and MS detection as well as by NMR spectroscopy. Subsequently, a series of site-directed mutants were generated with the aim of improving enzyme activity and investigating the importance of particular residues in determining substrate selectivity. The results reveal that the Phe-to-His mutants, i.e., OsaPAL2F134H and OsaPAL62F128H, exhibited higher TAL activity than the corresponding wild types, providing direct evidence that the Phe residue is responsible for substrate specificity. Mutagenesis studies also demonstrated that the Thr-to-Ser mutants, i.e., OsaPAL2T196S and OsaPAL62T194S, showed significantly higher substrate affinity than the wild types. Furthermore, the Gly-to-Ala mutants, i.e., OsaPAL2G209A and OsaPAL62G207A, showed higher PAL and TAL activities. These findings provide further insight into the genes responsible for OSW-1 biosynthesis and will facilitate the future application of OsaPALs in synthetic biology.
1. Introduction
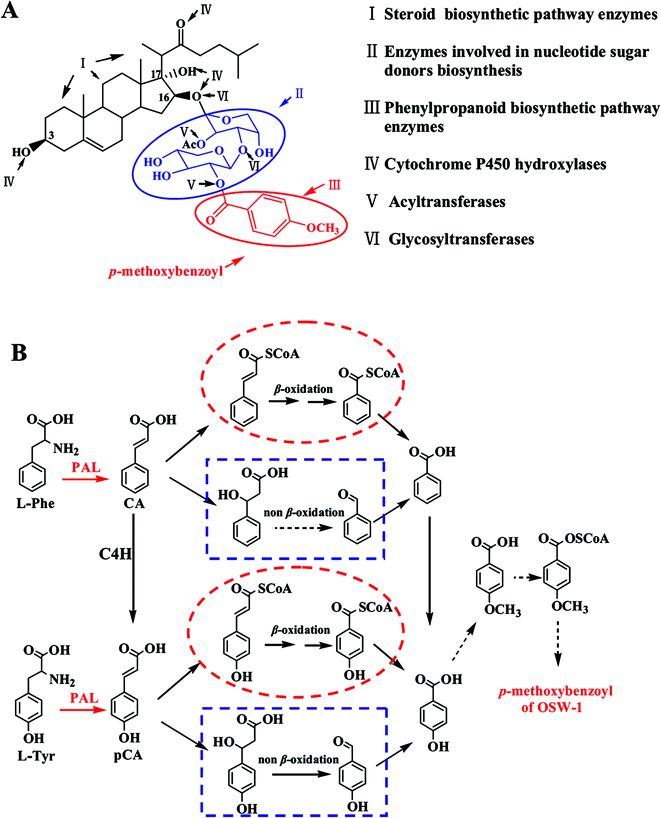
Phenylalanine ammonia lyase (PAL, EC 4.3.1.24), a member of the superfamily of ammonia lyases, catalyzes the conversion of L-phenylalanine to trans-cinnamic acid by a non-oxidative deamination. In recent years, considerable attention has been paid to the presence of PALs in medicinal plants since they are involved in the biosynthesis of the majority of phenol-containing secondary metabolites, many of which are pharmacologically active.1,2OSW-1 (Fig. 1A), which is a natural saponin isolated from Ornithogalum saundersiae, exhibits exceptionally potent antitumor activity both in vitro and in vivo.3–5 As such, it is a promising lead compound for the development of novel antitumor drugs; however, because of its low content in the plant and a long and laborious synthesis,6–10 little progress has been made in developing OSW-1 as a potential drug candidate after its discovery in 1992.11 Accordingly, we have initiated a project to identify an alternative, preparative-scale method to produce OSW-1 and to elucidate its biosynthetic pathway and the enzymes involved.
OSW-1 is characterized by a disaccharide moiety attached to the C-16 position of the steroid aglycone, which contains a p-methoxybenzoyl (MBz) and an acetyl (Ac) group.11 According to previous structure–activity relationship (SAR) studies, the disaccharide moiety is important for the cytotoxicity of OSW-1 and, in particular, the removal of the Ac and MBz groups decreases the activity of OSW-1 by approximately 1000-fold.3,12,13 Many previous studies have indicated that the p-hydroxybenzoic acid group is derived either from L-Phe or L-Tyr (Fig. 1B) through the respective PAL-catalyzed formation of trans-cinnamic acid or p-coumaric acid.14–16 Thus, it is likely that PALs play an important role in this phenylpropanoid biosynthetic pathway for OSW-1; however, to date, none of the PALs in O. saundersiae have been cloned or characterized.
PAL was first purified from Hordeum vulgare in 1961,17 and it was later found to be widespread in plants,18–20 fungi21–23 and prokaryotes.24–26 In plants, PALs are fairly ubiquitous; moreover, they can be found in monocots,27–30 dicots,1,18,19,31 gymnosperms,32–35 ferns,36 lycopods,36 liverworts,36 and algae;37 however, there are no reports about PALs in the Asparagaceae species.
Here, a gene family containing two OsaPAL genes was isolated for the first time from an Asparagaceae plant, O. saundersiae. After a successful functional characterization, a series of site-directed mutants were generated with the aim of improving enzyme activity and investigating the importance of specific residues in determining substrate selectivity. The in vitro assays indicated that we successfully improved both the PAL and TAL deamination activities of OsaPALs using a single amino acid substitution, a discovery that we believe will provide additional ways to improve PAL activity of other such enzymes. We also believe that a greater understanding of how OsaPALs participate in the biosynthesis of OSW-1 will facilitate future applications of OsaPALs in synthetic biology.
2. Experimental methods
2.1. Substrates, chemicals and enzymes
Materials (suppliers) used in this study were as follows: L-Phe and L-Tyr were used in the enzyme assays (Sigma-Aldrich Co. Ltd., St. Louis, MO, USA); In-Fusion® HD Cloning Kit, restriction enzymes (Takara Shuzo Co. Ltd., Kyoto, Japan) and Super RT cDNA Kit were used to synthesize full-length cDNAs and KOD-Plus-Neo DNA polymerases (Toyobo Co. Ltd., Osaka, Japan); RNeasy Plant Mini Kit was used for RNA extraction (Qiagen, Dusseldorf, Germany); Ni-Sepharose (Invitrogen, Carlsbad, CA, USA) and Fast Mutagenesis System kit were used for the site-directed mutagenesis (TransGen Biotech Co. Ltd., Beijing, China). All other chemicals used in this study were of analytical grade.2.2. Strains and plasmids
Prokaryotic expression vector, i.e. pET-28a (+), which was used for heterologous expression, was purchased from Novagen, Madison, USA. Expression host strain Transetta (DE3) and the cloning vector pEASY®-Blunt were purchased from TransGen Biotech Co. Ltd., Beijing, China. TG1 was used as a bacterial host for recombinant plasmid amplification. The host was grown in Luria–Bertani medium (10 g l−1 Bacto-Tryptone, 5 g l−1 Bacto-yeast extract, 10 g l−1 NaCl) or induced in TB medium (12 g l−1 Bacto-Tryptone, 24 g l−1 Bacto-yeast extract, 4 ml l−1 glycerol, 72 mM K2HPO4, 17 mM KH2PO4) supplemented with appropriate antibiotics for selection.2.3. Plant materials
O. saundersiae was grown under sterile conditions on 6,7-V medium38 at a temperature of 22 °C under a 16 h light/8 h dark cycle. Sterile bulbs were collected and used immediately for RNA isolation.2.4. Transcriptome sequencing and analysis
RNA extraction and cDNA library construction were performed as described in Kong et al.39 The resultant cDNA library was sequenced using Illumina HiSeq™ 2000. Short nucleotide reads obtained via Illumina sequencing were assembled using the Trinity software (http://www.trinity-software.com) to produce error-free, unique contiguous sequences (contigs). These contigs were ligated to obtain non-redundant unigenes, which could not be extended on either end. Unigene sequences were aligned by Blast X to protein databases like NCBI nr, Swiss-Prot, KEGG and COG (e-value < 0.00001), and aligned by Blast N to nucleotide databases nt (e-value < 0.00001), resulting in proteins with the highest sequence similarity with the given unigenes along with their functional annotations.2.5. Generation of full-length OsaPALs cDNA and sequence analysis
Because the assembled sequences were products of de novo assemblies, they were considered prone to error. To confirm whether a sequence represented a true gene product, experimental verification was performed by designing gene-specific primers (Table 1) for the OsaPAL full-length sequences, and then verifying the identity of amplified products by sequencing.| Primers | Sequences (5′–3′) | Description |
|---|---|---|
| FPAL1 | 5′-CATCATAATCTGACGGTTTTC-3′ | Forward primer used for OsaPAL2 amplification in the first round |
| FPAL2 | 5′-ATGGAGAACGGCAACGGTAA-3′ | Forward primer used for OsaPAL2 amplification in the second round |
| RPAL3 | 5′-CAGAATTATGAAATTCCAGCC-3′ | Reverse primer used for OsaPAL2 amplification in the first round |
| RPAL4 | 5′-TCAACATATTGGCAGCGGTGC-3′ | Reverse primer used for OsaPAL2 amplification in the second round |
| FET28aPAL2 | 5′-TCGCGGATCCGAATTCATGGAGAACGGCAACGGTAAC-3′ | Forward primer used for pET28a–OsaPAL2 construction |
| RET28aPAL2 | 5′-GTGCGGCCGCAAGCTTTCAACATATTGGCAGCGGTGC-3′ | Reverse primer used for pET28a–OsaPAL2 construction |
| FPAL5 | 5′-CAATCAGCCGTTTACGAGACC-3′ | Forward primer used for OsaPAL62 amplification in the first round |
| FPAL6 | 5′-ATGGAATCCCTCCACGCCAAC-3′ | Forward primer used for OsaPAL62 amplification in the second round |
| RPAL1 | 5′-CCGAAGTACTGAATGAAAATC-3′ | Reverse primer used for OsaPAL62 amplification in the first round |
| RPAL2 | 5′-CTAGCAAATGGGCAGGGGAG-3′ | Reverse primer used for OsaPAL62 amplification in the second round |
| FET28aPAL62 | 5′-TCGCGGATCCGAATTCATGGAATCCCTCCACGCCAAC-3′ | Forward primer used for pET28a–OsaPAL62 construction |
| RET28aPAL62 | 5′-GTGCGGCCGCAAGCTTCTAGCAAATGGGCAGGGGAG-3′ | Reverse primer used for pET28a–OsaPAL62 construction |
Total RNA isolated from sterile bulb tissue of O. saundersiae using an RNeasy Plant Mini Kit (Qiagen) was used as a template for reverse transcription with primer oligo (dT)20 primers and reverse transcriptase ReverTra Ace (TOYOBO) according to the manufacturer's instructions. The amplification of OsaPAL cDNAs was performed by a nested PCR method using KOD Plus Taq polymerase and gene-specific primers (Table 1). The amplified full-length cDNAs, i.e., OsaPAL2 and OsaPAL62, were each inserted into the pEASY®-Blunt vector to generate pEASY-OsaPAL2 and pEASY-OsaPAL62, respectively, for sequencing.
OsaPAL2 and OsaPAL62 were analyzed using online bioinformatic tools from NCBI and ExPASy. Open reading frame (ORF) finding was performed using the on-line program (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi). Various physicochemical parameters of proteins were evaluated using the ProtParam tool (http://web.expasy.org/protparam/). TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the transmembrane helices of proteins, and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was used to predict cleavage sites of signal peptides. Protein subcellular locations were predicted using TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP/), and functional sites of proteins were analyzed with PROSITE tools (http://prosite.expasy.org/). Concord (http://helios.princeton.edu/CONCORD/)40 was used to predict the secondary structures of OsaPAL2 and OsaPAL62, and protein multiple sequence alignment was performed using ClustalX (version 2.1).41 A phylogenetic tree was constructed using the neighbor-joining method with the MEGA5.1 program.42
2.6. Protein expression
The prokaryotic expression vector pET-28a (+) was digested with restriction endonucleases BamHI and EcoRI to generate the linearized vector. Plasmids pEASY-OsaPAL2 and pEASY-OsaPAL62 were used as templates for sub-cloning the full-length sequences of OsaPALs with respective specific primer pairs (Table 1). After gel purification, the PCR product was sub-cloned into pET-28a (+) according to the In-Fusion® HD Cloning Kit protocol. Finally, sequencing was used to verify the integrity of plasmids pET28a–OsaPAL2 and pET28a–OsaPAL62 with both of them containing a His6 tag.The expression plasmids pET28a–OsaPAL2 and pET28a–OsaPAL62 were transformed into the expression host strain Transetta (DE3) grown in LB agar containing 170 μg ml−1 chloromycetin and 50 μg ml−1 kanamycin. After overnight incubation at 37 °C, a single clone of each was used to inoculate 20 ml of LB/chloromycetin/kanamycin at 37 °C, which was shaken at 200 rpm until the OD at 600 nm reached a value of 1.0, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 into 50 ml TB supplemented with 170 μg ml−1 chloromycetin and 50 μg ml−1 kanamycin, and then shaken at 200 rpm at 37 °C until the OD at 600 nm reached 0.8. Subsequently, 0.1 mM IPTG was added and after shaking at 150 rpm at 20 °C overnight, the induced cells were harvested by centrifugation (7500g, 2 min) at 4 °C. The pellets were then either stored at −80 °C or used directly.
50 into 50 ml TB supplemented with 170 μg ml−1 chloromycetin and 50 μg ml−1 kanamycin, and then shaken at 200 rpm at 37 °C until the OD at 600 nm reached 0.8. Subsequently, 0.1 mM IPTG was added and after shaking at 150 rpm at 20 °C overnight, the induced cells were harvested by centrifugation (7500g, 2 min) at 4 °C. The pellets were then either stored at −80 °C or used directly.
2.7. Enzyme purification
For enzyme purification, all steps were performed at 4 °C. First, E. coli cells were washed and re-suspended in lysis buffer (pH 8.0, 20 mM sodium phosphate containing 5 mM imidazole and 300 mM NaCl). Cells were then lysed with a high-pressure homogenizer (800 bar, 3 passes), after which 1 U per ml DNaseI was added, and then the homogenate was incubated at 4 °C for approximately 2 h. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min, the supernatant was passed through a 0.2 μm pore-size filter to remove E. coli cell debris and other contaminants, and then loaded onto a pre-equilibrated column containing Ni-NTA resin. The column was washed with washing buffers (pH 8.0, 20 mM sodium phosphate buffer containing 20–50 mM imidazole and 300 mM NaCl) to remove non-specifically bound proteins, after which an elution buffer (pH 8.0, 20 mM sodium phosphate containing 300 mM imidazole and 300 mM NaCl) was used to elute the His6-tagged protein.
000g for 15 min, the supernatant was passed through a 0.2 μm pore-size filter to remove E. coli cell debris and other contaminants, and then loaded onto a pre-equilibrated column containing Ni-NTA resin. The column was washed with washing buffers (pH 8.0, 20 mM sodium phosphate buffer containing 20–50 mM imidazole and 300 mM NaCl) to remove non-specifically bound proteins, after which an elution buffer (pH 8.0, 20 mM sodium phosphate containing 300 mM imidazole and 300 mM NaCl) was used to elute the His6-tagged protein.
To remove small molecules such as imidazole, dialysis was performed. A semipermeable membrane with a molecular weight cutoff of 30 kDa was selected and approximately 20 ml protein sample was dialyzed against 1 l dialysis buffer (pH 8.0, 10 mM sodium phosphate) for 4 h at 4 °C with four changes of dialysis buffer. Proteins were then dried in a vacuum freeze-dryer and stored at −80 °C until use.
2.8. Enzyme activity and analysis
Enzyme activity of recombinant purified OsaPALs was unambiguously determined by a combination of HPLC-UV, HPLC-MS and 1H and 13C NMR spectroscopies using L-Phe and L-Tyr as substrates. The reaction mixture (200 μl), containing 0.1 M CHES buffer (pH 9.5), 100 mM L-Phe or 10 mM L-Tyr and different amounts of purified protein, was incubated at 37 °C for 30 min, and then terminated by adding 200 μl chloroform. After centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min, the supernatant was filtered through an 0.2 μm pore-size filter and analyzed by HPLC-UV on a HITACHI LaCrom elite L-2000 HPLC system (HITACHI, Toyokawa, Japan) using a C18 column [YMC-Pack ODS-A (5 μm, 12 nm, 250 × 4.6 mm)] and gradient elution using 0.05% aqueous trifluoroacetic acid (solvent A) and CH3CN (solvent B). After pre-equilibration in 98
000g for 10 min, the supernatant was filtered through an 0.2 μm pore-size filter and analyzed by HPLC-UV on a HITACHI LaCrom elite L-2000 HPLC system (HITACHI, Toyokawa, Japan) using a C18 column [YMC-Pack ODS-A (5 μm, 12 nm, 250 × 4.6 mm)] and gradient elution using 0.05% aqueous trifluoroacetic acid (solvent A) and CH3CN (solvent B). After pre-equilibration in 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 A
2 A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B (v/v), the sample was injected and chromatographed using a linear gradient to 35
B (v/v), the sample was injected and chromatographed using a linear gradient to 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 65 A
65 A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B (v/v) over 26 min at a flow rate of 1 ml min−1.
B (v/v) over 26 min at a flow rate of 1 ml min−1.
The reaction products of L-Phe (or L-Tyr), catalyzed by OsaPAL2 and OsaPAL62 [named OsaPAL2-L-Phe-p (or OsaPAL2-L-Tyr-p) and OsaPAL62-L-Phe-p (or OsaPAL62-L-Tyr-p), respectively], were isolated using a YMC semi-preparative column [YMC-Pack ODS-A (5 μm, 12 nm, 250 × 10 mm)] using a linear gradient elution from 40–65% solvent B in 15 min for L-Tyr products and a linear gradient elution from 40–80% solvent B in 15 min for L-Phe products, both of them at a flow rate of 2 ml min−1. UV detection at 275 and 310 nm was used for enzymatic products of L-Phe and L-Tyr, respectively. HPLC-MS was performed using an Agilent 1200 RRLC series HPLC system (Agilent Technologies, Waldbronn, Germany) coupled with a QTRAP tandem mass spectrometer (QTRAP 2000, Applied Biosystems/MDS SCIEX) equipped with a Turbo Ion spray source (Concord, ON, Canada) operated in the negative ionization mode and controlled by Analyst 1.5 software. Mass spectra were obtained in the enhanced full mass scan mode in the range of m/z 100–1000. NMR spectra of products dissolved in deuterated dimethyl sulfoxide (DMSO-d6) and placed in 5 mm NMR tubes were recorded using Bruker AVIII-600 and Bruker AVIII-500 NMR spectrometers (Bruker-Biospin, Germany), which were operated at 600 and 500 MHz, respectively. Chemical shifts (δ) and coupling constants (J) were provided in ppm and hertz (Hz), respectively.
2.9. Optimum pH of recombinant OsaPALs
To evaluate the effect of pH on enzyme activity, the following buffers were used: 0.1 M MES, pH 5.5–6.7; 0.1 M HEPES, pH 6.8–8.2; 0.1 M CHES, pH 8.6–10.0; 0.1 M CAPS, pH 9.7–11.1; and 0.1 M Na2HPO4–NaOH, pH 11.5–12.0. Assays were performed at a constant temperature of 37 °C for 20 min and monitored continuously with a multimode reader. Controls without enzymes were included and each experiment was performed in triplicate.2.10. Optimum temperature of recombinant OsaPALs
For determination of the optimal temperature, reactions were performed in 0.1 M CHES buffer (pH 9.5) and pre-incubated at different temperatures in the range 30–60 °C for 10 min. Substrates were added to initiate the reactions, and, after incubation for 15 min, during which time the change in absorption was measured, glacial acetic acid was added to terminate the reaction. Controls without enzymes were included and each experiment was performed in triplicate.2.11. Kinetic analysis of recombinant OsaPALs
Kinetic analysis of native enzymes was performed at pH 9.5 and 44 °C using a total volume of 200 μl, which contains various concentration ranges of substrates (L-Phe 9.76 μM to 20 mM; L-Tyr 15.6 μM to 8 mM) and purified proteins (for L-Phe: 6.3 μg OsaPAL2, 15.5 μg OsaPAL62, 47.32 μg OsaPAL2F134H, 36.5 μg OsaPAL62F128H, 19.6 μg OsaPAL2T196S, 22.8 μg OsaPAL62T194S, 23 μg OsaPAL2V202S, 647.2 μg OsaPAL62V200S, 8.8 μg OsaPAL2G209A, 12.1 μg OsaPAL62G207A; For L-Tyr: 135.6 μg OsaPAL2, 136.4 μg OsaPAL62, 236.6 μg OsaPAL2F134H, 182.84 μg OsaPAL62F128H, 175.2 μg OsaPAL2G209A, 120.6 μg OsaPAL62G207A). Assays for individual substrates were performed in triplicate for 10 min (L-Phe) or 20 min (L-Tyr). The formation of product was continuously monitored with a multimode reader. Kinetic constants values were determined from Lineweaver–Burk plots. All kinetic assays were performed in triplicate and controls without enzymes or substrates were included.2.12. Site-directed mutagenesis
Site-directed mutagenesis was used to investigate the importance of various amino acid residues in determining enzyme activity. Point and double mutations were introduced into OsaPALs by PCR-based amplification of the entire OsaPAL expression plasmid (pET28a–OsaPAL2 or pET28a–OsaPAL62) using two mutated oligonucleotide primers (Table 2), each complementary to the opposite strand of the vector. All components necessary for PCR-based mutagenesis were provided in the Fast Mutagenesis System kit and were used according to the manufacturer's instructions. All mutants were confirmed by sequencing and those plasmids with target substitutions and without other unwanted mutations were retained. Triple mutations were introduced in the same way using already existing mutants as templates.| Mutant | Template | Primer | Sequence |
|---|---|---|---|
| OsaPAL2F134H | pET28a–OsaPAL2 | PAL2F134HF | 5′-TGCAGAGAGAGTTGATAAGG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CTGAATGCCGG-3′ CTGAATGCCGG-3′ |
| PAL2F134HR | 5′-![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CCTTATCAACTCTCTCTGCAGGGCGCCCCC-3′ CCTTATCAACTCTCTCTGCAGGGCGCCCCC-3′ |
||
| OsaPAL2T196S | pET28a–OsaPAL2 | PAL2T196SF | 5′-TACGAGGCACCATC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GCCTCCGG-3′ GCCTCCGG-3′ |
| PAL2T196SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) GATGGTGCCTCGTAGAGGAAGGCA-3′ GATGGTGCCTCGTAGAGGAAGGCA-3′ |
||
| OsaPAL2V202S | pET28a–OsaPAL2 | PAL2V202SF | 5′-GCCTCCGGCGACCTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CCCTTGTCCT-3′ CCCTTGTCCT-3′ |
| PAL2V202SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TAGGTCGCCGGAGGCGGTGATGGTG-3′ TAGGTCGCCGGAGGCGGTGATGGTG-3′ |
||
| OsaPAL2G209A | pET28a–OsaPAL2 | PAL2G209AF | 5′-TTGTCCTACATTGCC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CTTCTCACCG-3′ CTTCTCACCG-3′ |
| PAL2G209AR | 5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GGCAATGTAGGACAAGGGGACTAGG-3′ GGCAATGTAGGACAAGGGGACTAGG-3′ |
||
| OsaPAL2V202S/G209A | pET28a–OsaPAL2 | PAL2G209AF | 5′-TTGTCCTACATTGCC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CTTCTCACCG-3′ CTTCTCACCG-3′ |
| PAL2DMR | 5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GGCAATGTAGGACAAGGGGC GGCAATGTAGGACAAGGGGC![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TAGG-3′ TAGG-3′ |
||
| OsaPAL2T196S/V202S/G209A | pET28a–OsaPAL2V202S/G209A | PAL2T196SF | 5′-TACGAGGCACCATC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GCCTCCGG-3′ GCCTCCGG-3′ |
| PAL2T196SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) GATGGTGCCTCGTAGAGGAAGGCA-3′ GATGGTGCCTCGTAGAGGAAGGCA-3′ |
||
| OsaPAL62F128H | pET28a–OsaPAL62 | PAL62F128HF | 5′-TTCAGAAGGAGCT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CAGACATCTCAACGCGGG-3′ CAGACATCTCAACGCGGG-3′ |
| PAL62F128HR | 5′-![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCTGATGAGCTCCTTCTGAAGGGCACCACC-3′ TCTGATGAGCTCCTTCTGAAGGGCACCACC-3′ |
||
| OsaPAL62T194S | pET28a–OsaPAL62 | PAL62T194SF | 5′-CTCCGCGGCACGATC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GCCTCCGGCG-3′ GCCTCCGGCG-3′ |
| PAL62T194SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) GATCGTGCCGCGGAGAGGGAGGCAC-3′ GATCGTGCCGCGGAGAGGGAGGCAC-3′ |
||
| OsaPAL62V200S | pET28a–OsaPAL62 | PAL62V200SF | 5′-GCCTCCGGCGACCTC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CCGTTGTCCT-3′ CCGTTGTCCT-3′ |
| PAL62V200SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) GAGGTCGCCGGAGGCGGTGATCGTG-3′ GAGGTCGCCGGAGGCGGTGATCGTG-3′ |
||
| OsaPAL62G207A | pET28a–OsaPAL62 | PAL62G207AF | 5′-ATATCGCC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ATCCTCACCG-3′ ATCCTCACCG-3′ |
| PAL62G207AR | 5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GGCGATATAGGACAACGG-3′ GGCGATATAGGACAACGG-3′ |
||
| OsaPAL62V200S/G207A | pET28a–OsaPAL62 | PAL62DMF | 5′-TTGTCCTATATCGCC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ATCCTCACCG-3′ ATCCTCACCG-3′ |
| PAL62DMR | 5′-CG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GGCGATATAGGACAACGGG GGCGATATAGGACAACGGG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AGG-3′ AGG-3′ |
||
| OsaPAL62T194S/V200S/G207A | pET28a–OsaPAL62V200S/G207A | PAL62T194SF | 5′-CTCCGCGGCACGATC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GCCTCCGGCG-3′ GCCTCCGGCG-3′ |
| PAL62T194SR | 5′-![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) GATCGTGCCGCGGAGAGGGAGGCAC-3′ GATCGTGCCGCGGAGAGGGAGGCAC-3′ |
2.13. Comparison of mutant activities
To compare the activities of wild-type and mutants, mutants were expressed and purified according to the methods described above. The purity of proteins was estimated by SDS-PAGE and pure proteins were dried in a vacuum freeze-dryer and stored at −80 °C. Prior to use, enzymes were dissolved in CHES buffer (0.1 M, pH 9.5), and the proteins were determined using the Bio-Rad protein assay (Bio-Rad, USA). HPLC was used to compare the catalytic activities of different mutants. Assays were performed in 1000 μl aqueous buffer containing 100 mM CHES (pH 9.5) and 25 μg enzyme. After pre-incubation at 37 °C for 10 min, substrates (50 mM L-Phe or 10 mM L-Tyr) were added and reactions run for 1, 2 or 3 h before being terminated by boiling for 5 min. Mixtures were then centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min) and 15 μl injected into the HPLC system. Each assay was performed in triplicate. Kinetic constants of some single mutants were also determined using the above procedure.
000g, 10 min) and 15 μl injected into the HPLC system. Each assay was performed in triplicate. Kinetic constants of some single mutants were also determined using the above procedure.
3. Results and discussion
3.1. Transcriptome analysis of OsaPAL homology
OSW-1 is a cholestane saponin featuring a novel 3β,16β,17α-trihydroxycholest-5-en-22-one aglycone with an acylated disaccharide attached to the 16-hydroxyl group (Fig. 1A). Biogenetic analysis showed that there were at least six kinds of enzymes responsible for OSW-1 biosynthesis, including terpenoid biosynthetic enzymes and steroid pathway enzymes, resulting in OSW-1 aglycone formation. Moreover, cytochrome P450 hydroxylase is able to add hydroxyl groups to the C-3, 16 and 17 positions of OSW-1 aglycone, and glycosyltransferase is involved in disaccharide moiety attachment to 16-OH of OSW-1 aglycone. Note that acyltransferases catalyze the introduction of the acetyl and the 4-methoxybenzoyl groups on the disaccharide moiety, while enzymes involved in nucleotide sugar donors biosynthesis provide glycosyl donors in glycosylation reactions and phenylpropanoid biosynthetic pathway enzymes convert aromatic amino acids to 4-methoxybenzoyl group (Fig. 1A). A total of more than 40 enzymes were deduced to be involved in the biosynthesis of OSW-1. It will take much more time to isolate and further characterize the function of all these genes using conventional molecular biology technologies. Thus, it is particularly important to apply a high-throughput method, which allows for drastically quicker and cheaper gene discovery and leads towards a far more comprehensive view of the OSW-1 biosynthetic pathway. The advent of next-generation sequencing approaches such as transcriptomic analysis provides a platform that has been proven to be critical in speeding up of the identification of a large number of related genes of secondary products. In the previous investigation, about 40 contigs and unigenes were retrieved and identified to be responsible for phenylpropanoid biosynthetic pathway from transcriptome sequence data of O. saundersiae.39 Furthermore, batch alignment results revealed that there are four unigenes showing high similarities with PALs. These four unigenes, namely, 25029, 25031, 26221 and 26880, were 2150, 2130, 1409, and 317 bp in length, respectively. A batch BlastX search of the GenBank with the four unigenes indicated that none of these unigenes contained a full-length CDS. Of them, unigenes 26880 and 25029 had the equal sequence identity with the same PALs deposited in GenBank, suggesting that they were situated within the same candidate PAL. It is the same case with unigene 26221 and 25031. Furthermore, sequence alignment analysis postulated that unigenes 26880 and 25029 were the 5′- and 3′-end of one candidate PAL, while unigenes 26221 and 25031 were the 5′ and 3′-end of another candidate PAL, respectively. Accordingly, a gene family containing two PAL cDNAs was acquired by transcriptomic analysis.3.2. Cloning and analysis of the full-length cDNAs encoding OsaPALs
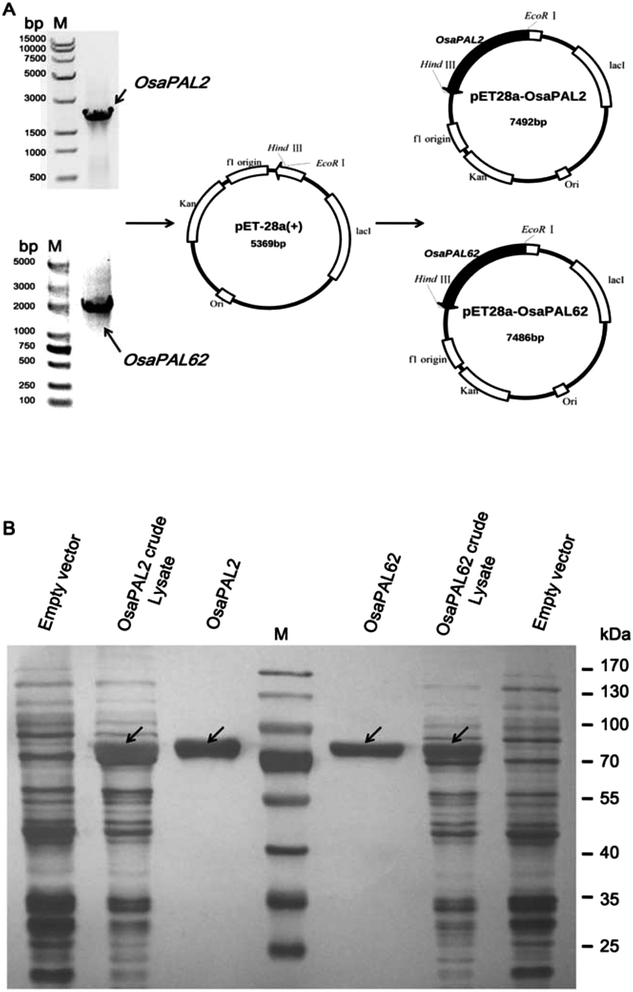
Two full-length members of the PAL gene family were isolated from O. saundersiae by nested PCR using gene-specific primers (Fig. 2A and Table 1). Identity between the cDNA sequences and the results of transcriptome sequencing, which were verified by sequencing, indicated the presence of a bona fide PAL gene family in plants. Sequence information for the two cDNAs, designated OsaPAL2 and OsaPAL62, was deposited in the GenBank database (OsaPAL2 accession number KF741222; OsaPAL62 accession number KF741223). OsaPAL2 was derived from the unigenes 26221 and 25031 and contained an ORF of 2136 bp. OsaPAL62 overlapped with unigenes 26880 and 25029 at the 5′ and 3′ ends, respectively, and it was 2130 bp in length. Sequencing identification proved the two cDNAs to be bona fide PALs of O. saundersiae, which is consistent with the fact that PALs are encoded by gene families in plant species such as Rubus idaeus,43Arabidopsis44 and Populus trichocarpa.45 Moreover, due to a lack of the genome sequence, we could not determine if the two cDNAs were derived from alternative splicing.The proteins encoded by OsaPAL2 and OsaPAL62 were predicted by ProtParam tool to be polypeptides containing 711 and 709 amino acids with molecular weights of 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103.9 and 76
103.9 and 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 570.6 Da, respectively. The instability indices (II) indicated that the two proteins were stable (II < 40), and their low grand average of hydropathicity (GRAVY) values of −0.139 (OsaPAL2) and −0.112 (OsaPAL62) indicate their hydrophobicity.46 The theoretical pI values of 5.64 (OsaPAL2) and 6.15 (OsaPAL62) were <7, revealing their acidic nature. The TMHMM tool indicated that OsaPAL2 or OsaPAL62 are not transmembrane proteins, and SignalP 4.1 showed no signal peptide or signal peptide cleavage sites, indicating that they are not secreted proteins. These results are consistent with the predictions of TargetP 1.1 that the two proteins are not located in chloroplasts, mitochondria or secretory pathways.
570.6 Da, respectively. The instability indices (II) indicated that the two proteins were stable (II < 40), and their low grand average of hydropathicity (GRAVY) values of −0.139 (OsaPAL2) and −0.112 (OsaPAL62) indicate their hydrophobicity.46 The theoretical pI values of 5.64 (OsaPAL2) and 6.15 (OsaPAL62) were <7, revealing their acidic nature. The TMHMM tool indicated that OsaPAL2 or OsaPAL62 are not transmembrane proteins, and SignalP 4.1 showed no signal peptide or signal peptide cleavage sites, indicating that they are not secreted proteins. These results are consistent with the predictions of TargetP 1.1 that the two proteins are not located in chloroplasts, mitochondria or secretory pathways.
According to the results provided by PROSITE tools, both OsaPALs contain a 17-amino acid motif (GTITASGDLVPLSYIAG), which is a conserved signature in the superfamily of ammonia lyases, including PAL, tyrosine ammonia lyase (TAL, EC 4.3.1.23) and histidine ammonia lyase (HAL, EC 4.3.1.3). The motif contains an active Ala–Ser–Gly (A–S–G) triad, which can autocatalytically be converted into a MIO (4-methylidene-imidazole-5-one) ring that serves as a prosthetic group and activates substrates by means of electrophilic interactions.47–49 With respect to the predicted secondary structure, both proteins had 8 β-sheets and 25 α-helices with the residues differing between the two proteins found primarily in random coils (Fig. 3).
3.3. Multiple sequence alignment and phylogenetic analysis
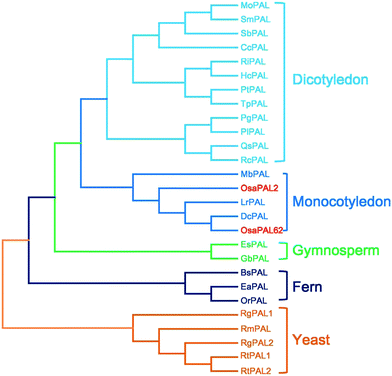
BlastP in NCBI (http://www.ncbi.nlmnih.gov/BLAST) and multi-alignment analysis according to CLUSTAL X algorithm indicate that the deduced polypeptide of OsaPAL2 is highly homologous to that of MbPAL (BAG70992.1) in Musa balbisiana. OsaPAL62 is highly homologous to PALs of some species, e.g., it shares 85% homology with the PAL sequence of Cinnamomum osmophloeum CoPAL (AFG26322.1), while the OsaPAL2 and OsaPAL62 sequences are 83.6% homologous.Phylogenetic analysis revealed that the PALs of angiosperm plants are divided into two clusters: those from dicotyledonous plants [e.g. PlPAL (AFI71896.1) from Paeonia lactiflora and RcPAL (AGH13333.1) from Rhus chinensis] and those from monocotyledon plants [e.g. MbPAL (BAG70992.1) from Musa balbisiana] (Fig. 4). Amino acid sequences of OsaPAL2 and OsaPAL62 belong to the latter cluster. It has been reported that PALs from monocotyledons always possess TAL activity,50,51 suggesting that OsaPAL2 and OsaPAL62 may have PAL/TAL activities. In the resulting phylogeny, OsaPAL62 and PALs from Lycoris radiate (LrPAL) and Dendrobium candidum (DcPAL) were clustered in the same clade, whereas OsaPAL2 was alone in a neighboring branch, suggesting that it belongs to a different subfamily. In addition, PALs of ferns and gymnosperms formed independent clusters, a result consistent with the evolution of plants. PALs of plants and yeasts have a common ancestor but are separated by a large evolutionary distance with only 36% homology between OsaPAL2 and RmPAL (CAA31486.1) from Rhodotorula mucilaginosa.
3.4. Expression of recombinant proteins and catalytic product analysis
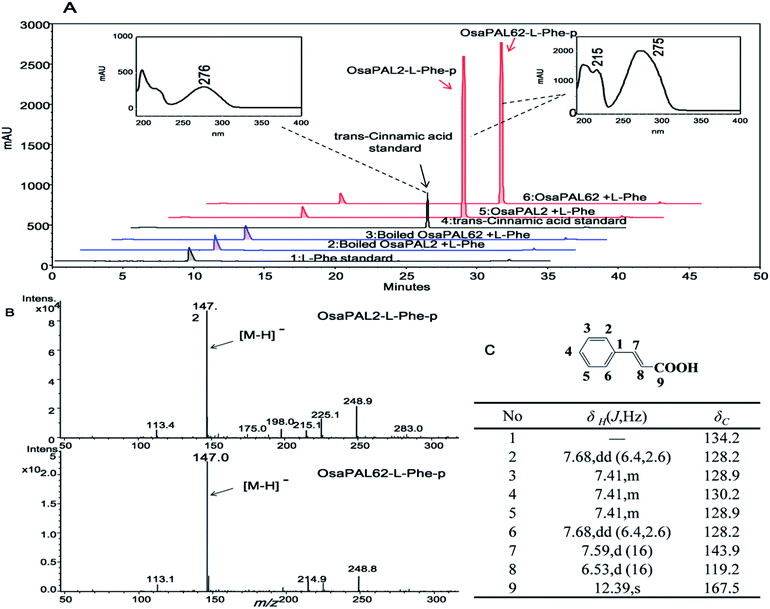
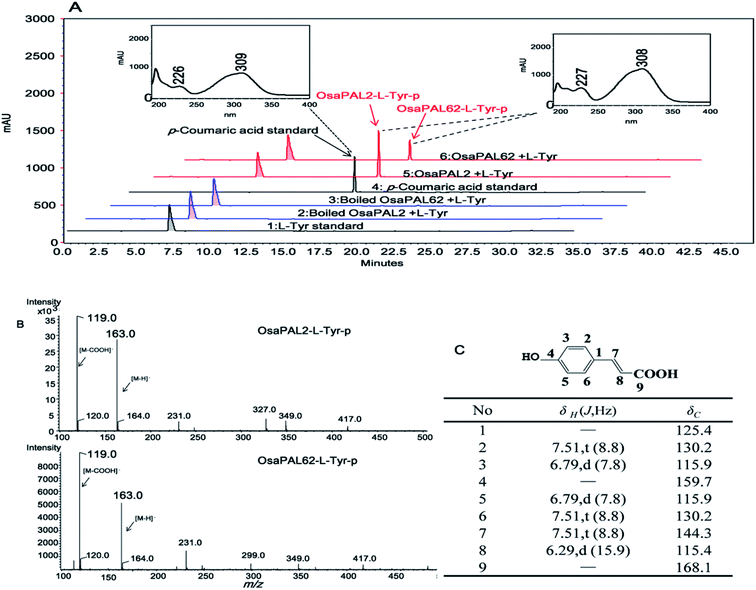
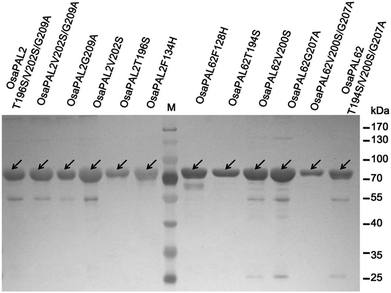
The two ORFs were sub-cloned into E. coli vector pET-28a (+) by the In-Fusion method, which resulted in the heterologous plasmids pET28a–OsaPAL2 and pET28a–OsaPAL62 (Fig. 2A). Recombinant His6-tagged OsaPAL2 and OsaPAL62 were expressed in E. coli and purified on Ni-NTA resin. SDS-PAGE analysis showed that both OsaPAL2 and OsaPAL62 were expressed and had molecular weights of approximately 75 kDa (Fig. 2B).Enzymes were incubated with L-Phe or L-Tyr, and the products were analyzed by HPLC-UV, HPLC-MS, and NMR. The data showed that the products of L-Phe catalyzed by OsaPAL2 and OsaPAL62 (OsaPAL2-L-Phe-p and OsaPAL62-L-Phe-p, respectively) had the same UV spectra as that of trans-cinnamic acid. In HPLC-MS, the most intense peaks of the two products were located at m/z 147 due to the loss of a hydrogen radical. The structure of these two products was further confirmed by NMR after semi-preparative isolation to be trans-cinnamic acid (Fig. 5).
For the corresponding products of L-Tyr, HPLC-UV analysis showed that both enzymes converted L-Tyr to the new products, OsaPAL2-L-Tyr-p and OsaPAL62-L-Tyr-p, which exhibit the same UV-spectra as that of p-coumaric acid. Both OsaPAL2-L-Tyr-p and OsaPAL62-L-Tyr-p exhibited an abundant ion at m/z 163 due to the loss of a hydrogen atom with subsequent loss of a carboxyl group to form an ion at m/z 119. NMR analysis further confirmed OsaPAL2-L-Tyr-p and OsaPAL62-L-Tyr-p to be p-coumaric acid (Fig. 6).
These results strongly suggest that OsaPAL2 and OsaPAL62 have both PAL and TAL activity, consistent with the activity of PALs present in other monocotyledonous plants such as Zea mays51 and Phyllostachys edulis.52
3.5. Biochemical analysis of OsaPALs
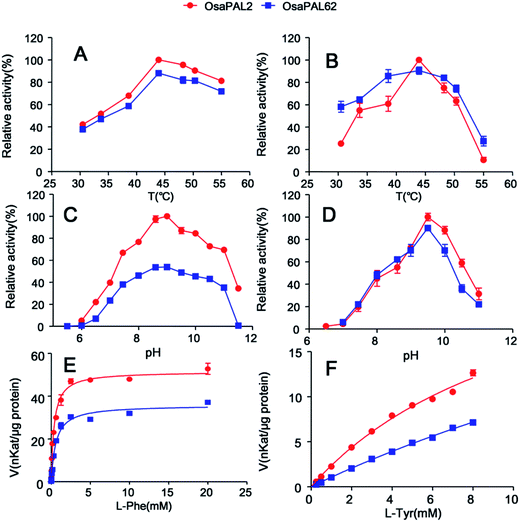
In many previous studies, the formation of trans-cinnamic acid was usually monitored at 275 or 280 nm.26,51,53 However, at this wavelength, the substrate L-Phe shows UV absorption (Fig. 5); thus, the determination of trans-cinnamic acid will be disturbed when using a multimode reader. Therefore, to eliminate the effect of the substrate, we monitored absorbance at 300 nm.The optimal pH and temperature for efficient enzyme activity of purified enzymes, OsaPAL2 and OsaPAL62, were found to be similar to those for L-Phe and L-Tyr deamination (Fig. 7). Both enzymes functioned better in the pH range of 8.5–10.0 and showed only low activity at pH <7.0 or >11.0. This optimum pH range is comparable to that of PALs isolated from other monocotyledonous species such as PePAL from Phyllostachys edulis (pH 8.5–9.0),52 SmPAL1 from Salvia miltiorrhiza (pH 8.7)2 and BoPAL4 from Bambusa oldhamii (pH 9.0).50 The effect of temperature on the activity of OsaPAL2 and OsaPAL62 was similar with both showing highest activity at 44 °C. Similar temperatures have been observed for maximum activity of PALs in Rhus chinensis (45 °C)1 and Arabidopsis (46–48 °C)44 with a slightly lower temperature being optimum for AvPAL in A. variabilis (40 °C)26 and a higher temperature being optimum for Pal in Helianthus annuus L. (55 °C).54
Assays were performed with L-Phe or L-Tyr as the substrate at an optimum pH and temperature to evaluate the kinetic constants of OsaPAL2 and OsaPAL62 (Fig. 7,8 and Table 3). Both wild-type enzymes displayed marked kinetic preference for L-Phe as Km of OsaPAL2 for L-Phe (371.5 ± 26.7 μM) was about 30-fold lower than that for L-Tyr (11690 ± 1410 μM) and kcat/Km for L-Phe (2160 M−1 s−1) was substantially higher than that for L-Tyr (39.65 M−1 s−1). A similar situation was found as a Km of OsaPAL62 for L-Phe (593.1 ± 48.6 μM) was about 60-fold lower than that for L-Tyr (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 ± 472 μM) and kcat/Km for L-Phe (934.9 M−1 s−1) was higher than that for L-Tyr (16.69 M−1 s−1). Assays performed to determine the kinetic constants of OsaPAL2 and OsaPAL62 for L-Tyr were unable to saturate the enzymes due to the lower appetency and solubility limitations of L-Tyr.44 The Km values of these enzymes for L-Phe were similar to those of other PALs such as the PAL of Oryza sativa (500 μM)55 and the BoPAL2 of B. oldhamii (333 μM).50 The kcat/Km values of these two wild-type enzymes for L-Phe were lower than those of the PALs in Zea mays (3.9 × 104 M−1 s−1)51 and Jatropha curcas L. (1.384 × 104 M−1 s−1).56
280 ± 472 μM) and kcat/Km for L-Phe (934.9 M−1 s−1) was higher than that for L-Tyr (16.69 M−1 s−1). Assays performed to determine the kinetic constants of OsaPAL2 and OsaPAL62 for L-Tyr were unable to saturate the enzymes due to the lower appetency and solubility limitations of L-Tyr.44 The Km values of these enzymes for L-Phe were similar to those of other PALs such as the PAL of Oryza sativa (500 μM)55 and the BoPAL2 of B. oldhamii (333 μM).50 The kcat/Km values of these two wild-type enzymes for L-Phe were lower than those of the PALs in Zea mays (3.9 × 104 M−1 s−1)51 and Jatropha curcas L. (1.384 × 104 M−1 s−1).56
| Substrate | Enzyme | Vmax (nkat per μg protein) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| L-Phe | OsaPAL2 | 51.48 ± 0.82 | 371.5 ± 26.7 | 0.8025 | 2160 |
| OsaPAL2F134H | 23.40 ± 0.23 | 1241 ± 44 | 0.3646 | 293.8 | |
| OsaPAL2T196S | 33.78 ± 0.36 | 662.9 ± 28.8 | 0.5264 | 794.1 | |
| OsaPAL2V202S | 12.21 ± 0.17 | 311.7 ± 21.1 | 0.1903 | 610.5 | |
| OsaPAL2G209A | 119.3 ± 2.2 | 790.4 ± 57.54 | 1.861 | 2354 | |
| OsaPAL62 | 35.82 ± 0.70 | 593.1 ± 48.6 | 0.5545 | 934.9 | |
| OsaPAL62F128H | 21.5 ± 0.2 | 1679 ± 43 | 0.3328 | 198.2 | |
| OsaPAL62T194S | 52.27 ± 0.46 | 1236 ± 39 | 0.8105 | 655.7 | |
| OsaPAL62V200S | 0.08034 ± 0.00130 | 118.9 ± 10.4 | 0.0003105 | 2.612 | |
| OsaPAL62G207A | 60.26 ± 0.61 | 529.2 ± 23.0 | 0.9332 | 1763 | |
| L-Tyr | OsaPAL2 | 29.73 ± 2.41 | 11690 ± 1410 | 0.4635 | 39.65 |
| OsaPAL2F134H | 12.35 ± 0.23 | 691.0 ± 46.0 | 0.1925 | 278.6 | |
| OsaPAL2G209A | 30.08 ± 1.33 | 3538 ± 365 | 0.4690 | 132.6 | |
| OsaPAL62 | 42.35 ± 4.40 | 39280 ± 472 | 0.6557 | 16.69 | |
| OsaPAL62F128H | 8.603 ± 0.161 | 1320 ± 74 | 0.1332 | 100.9 | |
| OsaPAL62G207A | 20.29 ± 1.56 | 9423 ± 1148 | 0.3143 | 33.35 |
The higher L-Phe and lower L-Tyr deamination activity of OsaPAL2 and OsaPAL62 is consistent with previous studies on PALs found in monocotyledons.51 Moreover, the OsaPALs were able to catalyze the reversible reaction whereby trans-cinnamic acid is converted to L-Phe, an ability that was also observed with some other PALs (data not shown).
3.6. Identification of specificity determining residues
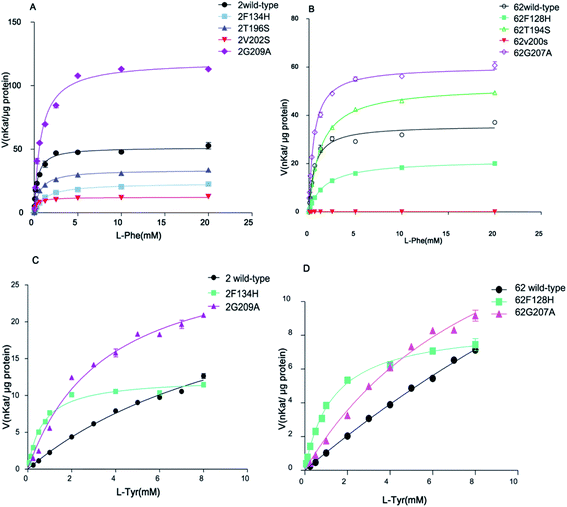
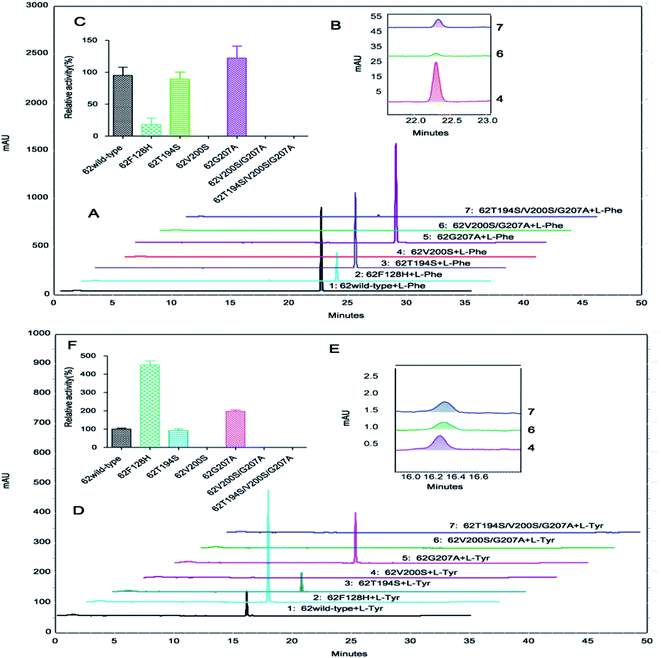
OsaPAL2 and OsaPAL62 also have TAL activity, allowing direct conversion of L-Tyr to p-coumarate, and thus eliminating the requirement for the hydroxylation step. This activity is valuable for designing heterologous pathways for producing target metabolites. One underlying mechanism of bi-functional PAL is based on the presence of a Phe residue controlling the substrate selection.57 Although several experiments have supported this hypothesis, various questions remain unaddressed. In a report by Watts et al.,57 when Phe144 residue was replaced by His in A. thaliana PAL1, the mutant displayed a marked reduction (80-fold) in kcat/Km value together with an 18-fold increase in catalytic efficiency towards tyrosine. This result clearly showed that Phe144 controlled substrate selection;57 however, Hsieh et al.50 did not obtain the same results. When Phe133 was substituted with His, only a slightly increased kcat/Km value toward tyrosine was observed in the F133H mutant. The authors inferred that other residues also contribute to the substrate selectivity of PALs. In view of this result, they proposed that further site-directed mutagenesis experiments would be necessary for elucidation of the underlying mechanism of substrate selectivity.50 This situation poses the question of how exactly the Phe residue functions in the substrate selectivity switch. Site-directed mutagenesis of Phe residue was accordingly performed in OsaPALs with the aim of identifying this function.TAL and PAL have different substrate specificities associated with a difference in a residue corresponding to F134 in OsaPAL2 and F128 in OsaPAL62 (Fig. 3). This residue gives rise to low L-Tyr deamination activity, which is improved by substituting the two residues with H to give OsaPAL2F134H and OsaPAL62F128H (Table 3, Fig. 8–10). The optimum pH and temperature for the two mutants are nearly the same as those of the wild-type proteins (ESI Fig. 1–3†). The kinetic parameters, however, varied markedly when F was mutated to H. For OsaPAL2F134H, the values of Km and kcat/Km for L-Tyr were 16.9-fold lower and 7.0-fold higher, respectively, than those for the wild-type enzyme. After F → H mutation, the activity toward L-Phe decreased with Km being 3-fold higher than that for OsaPAL2 and kcat/Km being reduced by 86% (Table 3). OsaPAL62F128H exhibited remarkable TAL activity compared with the wild-type enzyme (Table 3, Fig. 8 and 10) with values of Km and kcat/Km for L-Tyr were approximately 30-fold lower and 6-fold higher, respectively, than those for the wild-type enzyme. The substitution of F with H resulted in a significantly reduced activity for L-Phe with lower affinity (Km 2.8-fold higher than that of wild-type) and lower catalytic efficiency (kcat/Km 4.7-fold lower than that of the wild-type) (Table 3).
The fact that TAL activity of the OsaPALs is significantly increased in the F → H mutations proves that this F residue is associated with substrate specificity as has been reported in some previous studies.1,58 The compound, MIO, shown to be present in the crystal structure of PAL,59 is believed to be involved in the PAL reaction as a cofactor attacking the carbon atom at the C2 position of the aromatic ring of L-Phe in a Friedel–Crafts-type reaction.47 H134 and H128 of OsaPAL2F134H and OsaPAL62F128H, respectively, interact with the hydroxyl group of L-Tyr, resulting in an increase in affinity.58 The hydroxyl group of L-Tyr also acts as an electron-donating group to increase the electron density at the C1, C3 and C5 positions of the aromatic ring,60 thereby interfering with the attack by MIO; thus, the mutants retained higher catalytic activities towards L-Phe.61
3.7. Residue substitution successfully improves enzymes activity
The enzyme-kinetic properties of the purified proteins verified their biochemical function as bona fide OsaPALs, a finding which is expected to clarify the biosynthetic pathway of OSW-1. In addition, the successful characterization of the OsaPALs provides useful information for reconstructing the OSW-1 biosynthetic pathway and allows the scale-up of the production of OSW-1 using more active enzymes.Following the successful functional characterization, the two sequences were aligned with three known PALs from yeasts together with other three PALs from plants (Fig. 11). The reason for selection of these PALs was that PALs from yeasts61–63 appear to have higher catalytic activity than those from plants.1,51,64 Examination of the sequence alignment revealed three residues around the ASG triad that were not absolutely conserved. The three amino acids are T, V, and G in plant PALs, whereas, in yeast PALs, the equivalent residues were S, S, and A. This is an interesting phenomenon that has not been reported to date in the literature. To identify the function of the three amino acids, site-directed mutagenesis was performed based on the consensus approach.65–67
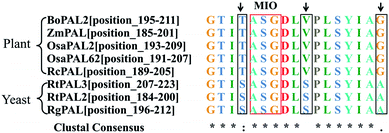 | ||
| Fig. 11 Multiple alignments of PAL signatures of plants and yeasts. The red box highlights the active site motif (A–S–G), which undergoes autocatalysis to form MIO. The residues that differ in yeasts and plants are shown in black boxes. The sequences used are OsaPAL2, OsaPAL62, Rhus chinensis RcPAL (AGH13333), Zea mays ZmPAL (NP_001105334), Bambusa oldhamii BoPAL2 (ACN62413.1), Rhodotorula glutinis JN-1 RgPAL (see ref. 61), Rhodosporidium toruloides RtPAL3 (P11544), Rhodosporidium toruloides RtPAL2 (AAA33883.1). | ||
The mutants were OsaPAL2T196S and OsaPAL62T194S, in which T196 of OsaPAL2 and T194 of OsaPAL62 were replaced with S; OsaPAL2V202S and OsaPAL62V200S, in which V202 of OsaPAL2 and V200 of OsaPAL62 were replaced with S; and OsaPAL2G209A and OsaPAL62G207A, in which G209 of OsaPAL2 and G207 of OsaPAL62 were replaced with A. In addition, two double mutants, OsaPAL2V202S/G209A and OsaPAL62V200S/G207A, and two triple mutants, OsaPAL2T196S/V202S/G209A and OsaPAL62T194S/V200S/G207A, were constructed.
After protein expression and purification (Fig. 12), the optimum pH and temperature for these mutants were determined to be the same as those for the wild proteins (ESI Fig. 1–3†). Then, the catalytic activities of these mutants were evaluated (Table 3 and Fig. 8–10), which reveal that although all the mutations lay near the PAL motif, their effects were diverse (Table 3). Using L-Phe as a substrate, both T → S mutations (OsaPAL2T196S and OsaPAL62T194S) and V → S mutations (OsaPAL2V202S and OsaPAL62V200S) exhibited lower catalytic efficiency with values of kcat/Km lower than those of the wild-type enzymes, in particular of OsaPAL62V200S whose kcat/Km value was only 0.33% of that of OsaPAL62.
Interestingly, the T → S mutation, where one hydrophilic amino acid replaces another, showed lower affinity with Km values 2-fold higher and lower catalytic efficiency compared to the wild-type. In contrast, the V → S mutation, where a hydrophilic amino acid replaces a hydrophobic one, showed higher affinity than the wild-type. Furthermore, the G → A mutants were did not exhibit significant difference in affinity; however, they exhibited higher catalytic efficiency than the wild type.
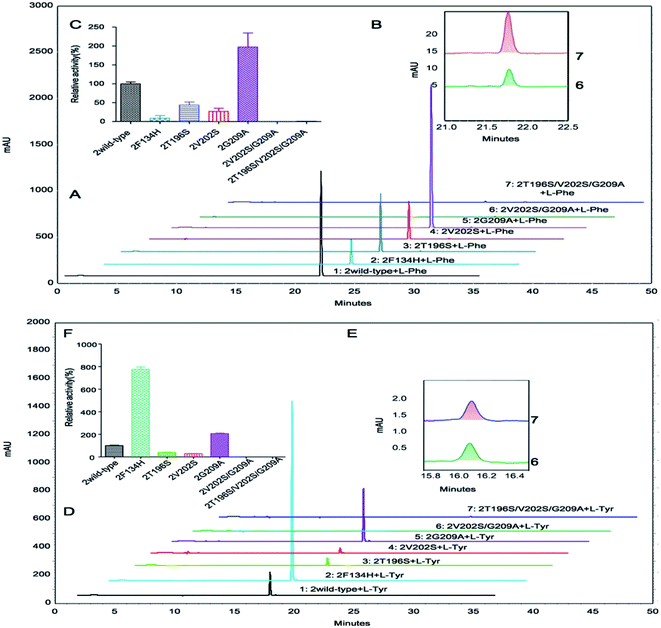
The double mutants OsaPAL2V202S/G209A and OsaPAL62V200S/G207A exhibited activities too low to be detected by the multimode reader; however, by HPLC analysis, they were shown to have activities that were only 0.006% and 0.129%, respectively, of the wild-type activities for L-Phe (Fig. 9 and 10). The introduction of a third mutation to afford the triple mutants OsaPAL2T196S/V202S/G209A and OsaPAL62T194S/V200S/G207A restored some degree of activity for L-Phe (0.016–0.339%) (Fig. 9 and 10). The relative activities of other mutants were also evaluated by HPLC and shown to be consistent with their kinetic constants. In brief, the activity of OsaPAL2G209A and OsaPAL62G207A was highest, followed by the wild-type, followed by OsaPAL2T196S and OsaPAL62T194S with slightly lower activity than the wild-type, and finally OsaPAL2V202S and OsaPAL62V200S. Activities of the double mutants OsaPAL2V202S/G209A and OsaPAL62V200S/G207A and triple mutants OsaPAL2T196S/V202S/G209A and OsaPAL62T194S/V200S/G207A were negligible. Similar results were obtained by HPLC for L-Tyr deamination activity (Fig. 9 and 10).
In both of the two proposed reaction mechanisms for PAL,48,60,68,69 the A–S–G triad in the motif can rearrange to a MIO ring to attack substrates. The importance of the triad has been demonstrated in many studies but little research has focused on other conserved amino acids around it.
The results described above indicate that residues adjacent to the A–S–G triad may play different roles. In considering the Km values of single mutants in relation to the positions of mutations, the higher affinity but lower catalytic efficiency of OsaPAL2V202S and OsaPAL62V200S suggests that the S202 in the former and S200 in the latter may form hydrogen bonds with the β-proton of the substrate and hinder its elimination. However, the elucidation of the exact mechanism needs further investigation.
4. Conclusions
4.1. First functional characterization of PAL cDNAs from Asparagaceae plant species
PALs are fairly ubiquitous in plants, including monocots, dicots, gymnosperms, ferns, lycopods, liverworts, and algae; however, there are no reports about PALs from Asparagaceae plant species.4.2. The function of three amino acids around ASG triad
PALs from yeasts appear to have a higher catalytic activity than those from plants. The examination of the sequence alignment revealed three residues around the ASG triad that were not absolutely conserved. The three amino acids are T, V, and G in the plant PALs, whereas in yeast PALs, the equivalent residues were S, S, and A. The amino acid difference may be the reason for the discrepancy in catalytic activity between plant PALs and yeast PALs. Mutagenesis studies demonstrated that the Thr-to-Ser mutants, OsaPAL2T196S and OsaPAL62T194S showed significantly higher substrate affinity than the wild-types; moreover, the Gly-to-Ala mutants, OsaPAL2G209A and OsaPAL62G207A showed higher PAL and TAL activities. To the best of our knowledge, this is the first report about these three mutants. It is also the first time that we have improved both PAL and TAL activities by a single amino acid substitution.4.3. The function of phenylalanine as a substrate selective switch
There are some confusing results about the function of Phe as a substrate selective switch. In the paper presented by Watts et al.,58 when Phe144 residue was replaced by His in A. thaliana PAL1, the mutant displayed a marked reduction (80-fold) in kcat/Km value together with an 18-fold increase in the catalytic efficiency towards tyrosine. This result clearly showed that Phe144 controlled the substrate selection; however, this was not the case in the report published by Hsieh et al.51 When Phe133 was substituted with His, a slightly increased kcat/Km value toward tyrosine was found in the F133H mutant. They inferred that other residues also contribute to the substrate selectivity of PALs. To further clarify the function of phenylalanine, two mutants OsaPAL2F134H and OsaPAL62F128H were produced by site-directed mutagenesis. The results corroborated that Phe does indeed control substrate selection.Acknowledgements
This work was supported by National Mega-project for Innovative Drugs (no. 2012ZX09301002), Open Foundation of State Key Laboratory of Bioactive Substance and Function of Natural Medicines (B-2011-4) and PUMC Youth Fund (2012J21, 3332013112).References
- A. B. Oliveira, C. F. Moura, E. Gomes-Filho, C. A. Marco, L. Urban and M. R. Miranda, PLoS One, 2013, 8, e56354 CAS.
- J. Song and Z. Wang, Mol. Biol. Rep., 2009, 36, 939–952 CrossRef CAS PubMed.
- Y. Mimaki, M. Kuroda, A. Kameyama, Y. Sashida, T. Hirano, K. Oka, R. Maekawa, T. Wada, K. Sugita and J. A. Beutler, Bioorg. Med. Chem. Lett., 1997, 7, 633–636 CrossRef CAS.
- K. Tamura, H. Honda, Y. Mimaki, Y. Sashida and H. Kogo, Br. J. Pharmacol., 1997, 121, 1796–1802 CrossRef CAS PubMed.
- Y. Zhou, C. Garcia-Prieto, D. A. Carney, R. H. Xu, H. Pelicano, Y. Kang, W. Yu, C. Lou, S. Kondo, J. Liu, D. M. Harris, Z. Estrov, M. J. Keating, Z. Jin and P. Huang, J. Natl. Cancer Inst., 2005, 97, 1781–1785 CrossRef CAS PubMed.
- S. Deng, B. Yu, Y. Lou and Y. Hui, J. Org. Chem., 1999, 64, 202–208 CrossRef CAS PubMed.
- J. W. Morzycki and A. Wojtkielewicz, Carbohydr. Res., 2002, 337, 1269–1274 CrossRef CAS.
- M. Tsubuki, S. Matsuo and T. Honda, Tetrahedron Lett., 2008, 49, 229–232 CrossRef CAS PubMed.
- W. Yu and Z. Jin, J. Am. Chem. Soc., 2002, 124, 6576–6583 CrossRef CAS PubMed.
- J. Xue, P. Liu, Y. Pan and Z. Guo, J. Org. Chem., 2008, 73, 157–161 CrossRef CAS PubMed.
- S. Kubo, Y. Mimaki, M. Terao, Y. Sashida, T. Nikaido and T. Ohmoto, Phytochemistry, 1992, 31, 3969–3973 CrossRef CAS.
- M. Kuroda, Y. Mimaki, A. Yokosuka, F. Hasegawa and Y. Sashida, J. Nat. Prod., 2002, 65, 1417–1423 CrossRef CAS PubMed.
- M. Kuroda, Y. Mimaki, A. Yokosuka, Y. Sashida and J. A. Beutler, J. Nat. Prod., 2001, 64, 88–91 CrossRef CAS PubMed.
- A. Podstolski, D. Havkin-Frenkel, J. Malinowski, J. W. Blount, G. Kourteva and R. A. Dixon, Phytochemistry, 2002, 61, 611–620 CrossRef CAS.
- D. Sircar and A. Mitra, J. Plant Physiol., 2009, 166, 1370–1380 CrossRef CAS PubMed.
- R. Loscher and L. Heide, Plant Physiol., 1994, 106, 271–279 Search PubMed.
- J. Koukol and E. E. Conn, J. Biol. Chem., 1961, 236, 2692–2698 CAS.
- X. Hou, F. Shao, Y. Ma and S. Lu, Mol. Biol. Rep., 2013, 40, 4301–4310 CrossRef CAS PubMed.
- C. J. Dong and Q. M. Shang, Planta, 2013, 238, 35–49 CrossRef CAS PubMed.
- S. Giberti, C. M. Bertea, R. Narayana, M. E. Maffei and G. Forlani, J. Plant Physiol., 2012, 169, 249–254 CrossRef CAS PubMed.
- H. Yoon, Y. H. You, Y. E. Kim, Y. J. Kim, W. S. Kong and J. G. Kim, J. Microbiol. Biotechnol., 2013, 23, 1055–1059 CrossRef CAS PubMed.
- C. A. Vaslet, R. L. Strausberg, A. Sykes, A. Levy and D. Filpula, Nucleic Acids Res., 1988, 16, 11382 CrossRef CAS PubMed.
- C. W. Abell and R. S. Shen, Methods Enzymol., 1987, 142, 242–253 CAS.
- K. Kovacs, G. Banoczi, A. Varga, I. Szabo, A. Holczinger, G. Hornyanszky, I. Zagyva, C. Paizs, B. G. Vertessy and L. Poppe, PLoS One, 2014, 9, e85943 Search PubMed.
- J. S. Williams, M. Thomas and D. J. Clarke, Microbiology, 2005, 151, 2543–2550 CrossRef CAS PubMed.
- M. C. Moffitt, G. V. Louie, M. E. Bowman, J. Pence, J. P. Noel and B. S. Moore, Biochemistry, 2007, 46, 1004–1012 CrossRef CAS PubMed.
- X. H. Wang, M. Gong, L. Tang, S. Zheng, J. D. Lou, L. Ou, J. Gomes-Laranjo and C. Zhang, Mol. Biol. Rep., 2013, 40, 97–107 CrossRef PubMed.
- Q. Jin, Y. Yao, Y. Cai and Y. Lin, PLoS One, 2013, 8, e62352 CAS.
- Y. Jiang, B. Xia, L. Liang, X. Li, S. Xu, F. Peng and R. Wang, Mol. Biol. Rep., 2013, 40, 2293–2300 CrossRef CAS PubMed.
- C. Fang, Y. Zhuang, T. Xu, Y. Li, Y. Li and W. Lin, J. Chem. Ecol., 2013, 39, 204–212 CrossRef CAS PubMed.
- F. Xu, G. Deng, S. Cheng, W. Zhang, X. Huang, L. Li, H. Cheng, X. Rong and J. Li, Molecules, 2012, 17, 7810–7823 CrossRef CAS PubMed.
- U. R. Bagal, J. H. Leebens-Mack, W. W. Lorenz and J. F. Dean, BMC Genomics, 2012, 13, 1–9 CrossRef PubMed.
- W. H. Xiao, J. S. Cheng and Y. J. Yuan, J. Biotechnol., 2009, 139, 222–228 CrossRef CAS PubMed.
- R. W. Whetten and R. R. Sederoff, Plant Physiol., 1992, 98, 380–386 CrossRef CAS PubMed.
- G. Hao, X. Du, F. Zhao and H. Ji, Biotechnol. Lett., 2010, 32, 305–314 CrossRef CAS PubMed.
- M. R. Young, G. H. N. Towers and A. C. Neish, Can. J. Bot., 1966, 44, 341–349 CrossRef CAS.
- U. Czichi and H. Kindi, Hoppe-Seyler's Z. Physiol. Chem., 1975, 356, 475–485 CrossRef CAS.
- I. A. Veliky and S. M. Martin, Can. J. Microbiol., 1970, 16, 223–226 CrossRef CAS.
- J. Q. Kong, D. Lu and Z. B. Wang, Molecules, 2014, 19, 1608–1621 CrossRef CAS PubMed.
- Y. Wei, J. Thompson and C. Floudas, Proc. R. Soc. A, 2012, 468, 831–850 CrossRef CAS.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS PubMed.
- K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, Mol. Biol. Evol., 2011, 28, 2731–2739 CrossRef CAS PubMed.
- A. Kumar and B. E. Ellis, Plant Physiol., 2001, 127, 230–239 CrossRef CAS PubMed.
- F. C. Cochrane, L. B. Davin and N. G. Lewis, Phytochemistry, 2004, 65, 1557–1564 CrossRef CAS PubMed.
- R. Shi, C. M. Shuford, J. P. Wang, Y. H. Sun, Z. Yang, H. C. Chen, S. Tunlaya-Anukit, Q. Li, J. Liu, D. C. Muddiman, R. R. Sederoff and V. L. Chiang, Planta, 2013, 238, 487–497 CrossRef CAS PubMed.
- J. Kyte and R. F. Doolittle, J. Mol. Biol., 1982, 157, 105–132 CrossRef CAS.
- L. Poppe, Curr. Opin. Chem. Biol., 2001, 5, 512–524 CrossRef CAS.
- L. Poppe and J. Retey, Angew. Chem., Int. Ed., 2005, 44, 3668–3688 CrossRef CAS PubMed.
- J. Retey, Biochim. Biophys. Acta, 2003, 1647, 179–184 CrossRef CAS.
- L. S. Hsieh, G. J. Ma, C. C. Yang and P. D. Lee, Phytochemistry, 2010, 71, 1999–2009 CrossRef CAS PubMed.
- J. Rosler, F. Krekel, N. Amrhein and J. Schmid, Plant Physiol., 1997, 113, 175–179 CAS.
- Z. M. Gao, X. C. Wang, Z. H. Peng, B. Zheng and Q. Liu, Plant Cell Rep., 2012, 31, 1345–1356 CrossRef CAS PubMed.
- J. D. Cui, L. L. Li and H. J. Bian, PLoS One, 2013, 8, e80581 Search PubMed.
- J. Jorrin, R. Lopez-Valbuena and M. Tena, Biochem. Int., 1991, 24, 1–11 CAS.
- A. D. Sarma and R. Sharma, Phytochemistry, 1999, 50, 729–737 CrossRef CAS.
- J. Gao, S. Zhang, F. Cai, X. Zheng, N. Lin, X. Qin, Y. Ou, X. Gu, X. Zhu, Y. Xu and F. Chen, Mol. Biol. Rep., 2012, 39, 3443–3452 CrossRef CAS PubMed.
- K. T. Watts, B. N. Mijts, P. C. Lee, A. J. Manning and C. Schmidt-Dannert, Chem. Biol., 2006, 13, 1317–1326 CrossRef CAS PubMed.
- G. V. Louie, M. E. Bowman, M. C. Moffitt, T. J. Baiga, B. S. Moore and J. P. Noel, Chem. Biol., 2006, 13, 1327–1338 CrossRef CAS PubMed.
- J. C. Calabrese, D. B. Jordan, A. Boodhoo, S. Sariaslani and T. Vannelli, Biochemistry, 2004, 43, 11403–11416 CrossRef CAS PubMed.
- B. Schuster and J. Rétey, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 8433–8437 CrossRef CAS.
- L. Zhu, W. Cui, Y. Fang, Y. Liu, X. Gao and Z. Zhou, Biotechnol. Lett., 2013, 35, 751–756 CrossRef CAS PubMed.
- I. L. Bazukian, A. E. Vardanian, A. A. Ambartsumian, P. V. Tozalakian and G. Popov Iu, Prikl. Biokhim. Mikrobiol., 2009, 45, 23–27 CAS.
- C. T. Evans, K. Hanna, D. Conrad, W. Peterson and M. Misawa, Appl. Microbiol. Biotechnol., 1987, 25, 406–414 CAS.
- L. S. Hsieh, C. S. Yeh, H. C. Pan, C. Y. Cheng, C. C. Yang and P. D. Lee, Protein Expression Purif., 2010, 71, 224–230 CrossRef CAS PubMed.
- M. Lehmann and M. Wyss, Curr. Opin. Biotechnol., 2001, 12, 371–375 CrossRef CAS.
- R. A. Chica, N. Doucet and J. N. Pelletier, Curr. Opin. Biotechnol., 2005, 16, 378–384 CrossRef CAS PubMed.
- S. Lutz, Curr. Opin. Biotechnol., 2010, 21, 734–743 CrossRef CAS PubMed.
- K. R. Hanson and E. A. Havir, Arch. Biochem. Biophys., 1970, 141, 1–17 CrossRef CAS.
- J. D. Hermes, P. M. Weiss and W. W. Cleland, Biochemistry, 1985, 24, 2959–2967 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra03385j |
| This journal is © The Royal Society of Chemistry 2014 |