Single-step alcohol-free synthesis of core–shell nanoparticles of β-casein micelles and silica
Stef Kerkhofs†
a,
Frederic Leroux†b,
Lionel Allouchec,
Randy Mellaertsa,
Jasper Jammaera,
Alexander Aertsa,
Christine E. A. Kirschhocka,
Pieter C. M. M. Magusina,
Francis Taulellead,
Sara Balsb,
Gustaaf Van Tendeloob and
Johan A. Martens*a
aCentre for Surface Chemistry and Catalysis, KU Leuven, Heverlee, Belgium. E-mail: Johan.Martens@biw.kuleuven.be
bElectron Microscopy for Materials Science (EMAT), UAntwerp, Belgium
cService de R.M.N., Institut de Chimie, Université de Strasbourg, France
dTectospin, Institut Lavoisier, Université de Versailles St Quentin en Yvelines, France
First published on 28th May 2014
Abstract
A new, single-step protocol for wrapping individual nanosized β-casein micelles with silica is presented. This biomolecule-friendly synthesis proceeds at low protein concentration at almost neutral pH, and makes use of sodium silicate instead of the common silicon alkoxides. This way, formation of potentially protein-denaturizing alcohols can be avoided. The pH of the citrate-buffered synthesis medium is close to the isoelectric point of β-casein, which favours micelle formation. A limited amount of sodium silicate is added to the protein micelle suspension, to form a thin silica coating around the β-casein micelles. The size distribution of the resulting protein–silica structures was characterized using DLS and SAXS, as well as 1H NMR DOSY with a dedicated pulsed-field gradient cryo-probehead to cope with the low protein concentration. The degree of silica-condensation was investigated by 29Si MAS NMR, and the nanostructure was revealed by advanced electron microscopy techniques such as ESEM and HAADF-STEM. As indicated by the combined characterization results, a silica shell of 2 nm is formed around individual β-casein micelles giving rise to separate protein core–silica shell nanoparticles of 17 nm diameter. This alcohol-free method at mild temperature and pH is potentially suited for packing protein molecules into bio-compatible silica nanocapsules for a variety of applications in biosensing, therapeutic protein delivery and biocatalysis.
Introduction
Preservation of protein functionality under artificial conditions is a key issue in applications such as therapeutic protein delivery, biocatalysis and bio-sensing.1–9 Steric restriction in a confined nanospace is one of the successful approaches to enhance the conformational stability of a protein.3,5,9–13 In particular, the enclosure of a single or few protein molecules into polymer or silica capsules is an attractive means to expand the conditions under which proteins can be functionally applied.10,12–21 The protein nanocapsules can even be made responsive to selected triggering, depending on the intended application. For instance, organic polymer shells in therapeutic delivery systems can be engineered to degrade at a targeted pH.22 One type of encapsulation involves covalently attaching functional groups to the protein surface, followed by a polymerization of organic22 or inorganic10,14 monomers forming a shell around the protein. For more fragile enzymes, however, such encapsulation methods involving chemical modification may cause a loss of functionality.10Silica is an attractive wrapping agent because of its limited solubility under physiological conditions, flexibility in handling and biocompatibility.1,2,23–26 Encapsulation of individual living cells in silica gels has been reported, including prokaryotes, yeast cells, plant and animal cells.2–4,25,27–33 The encapsulation of yeast cells with silica has been demonstrated to extend their lifespan, and to protect the cells from unfavourable external conditions.31,32 Concerning biomolecules, many delicate water-soluble and membrane-bound proteins have been successfully immobilized in large silica gel bodies.2–4,7,8,26,34
Encapsulation of individual proteins in silica capsules has also been achieved.14,15,19,35,36 One approach involves the use of triblock copolymer, organic solvent and silicon alkoxide. A protein such as met-myoglobin was solubilized in cyclohexane by incorporation in poly(ethylene oxide)–poly(propylene oxide)–poly(ethylene oxide) triblock copolymer (EO20–PO70–EO20, P123, Pluronic®) conjugates. The added tetramethyl orthosilicate (TMOS) was hydrolyzed in the presence of traces of water, and polymerized around the nano-emulsion of met-myoglobin/P123 conjugates. These silica-wrapped protein capsules were precipitated, dried and suspended in aqueous solution.19 Besides the complexity of this synthesis protocol, including solvent removal steps, a general disadvantage of using organic solvents in the encapsulation process is the risk of protein denaturation. Nevertheless, in the reported cases, proteins entrapped inside silica capsules retained their functionality, secondary structure and the desired improvement of protein stability.10,14,19
Typically, silica polymerization around proteins is achieved by hydrolysis and condensation of silicon alkoxides.1,37,38 However, hydrolysis of alkoxides like TMOS and TEOS releases stoichiometric quantities of methanol and ethanol, which may cause protein denaturation.1,38 To avoid alcohol formation the use of sodium silicate as a silica source has been suggested.2,38 In this way, proteins were successfully embedded into bulk silica matrices.38,39
Our new approach for synthesis of protein–silica core–shell nanoparticles was inspired by the formation process of COK-12. This ordered mesoporous silica material has pores which are formed using micelles of sacrificial triblock copolymer as supramolecular template.40,41 COK-12 formation proceeds at ambient temperature and quasi-neutral pH conditions in citrate buffer, and uses sodium silicate solution as silica source. This starting solution of sodium silicate is a mixture of monomer and silicate oligomers.42 In the first stage of the formation process, silica is precipitated on the individual spherical micelles of the P123 triblock copolymer. Provided the silica addition remains limited, a suspension of silica-decorated P123 micelles is obtained, stabilized by steric repulsion. Adding more silica leads to the supramolecular assembly of COK-12 material.42 Spherical P123 micelles measure ca. 14 nm in diameter, similar to the size of large proteins, or protein micelles.41,43–46 We adopted the first stage of the COK-12 formation process, in absence of the synthetic copolymer, for the single-step preparation of discrete protein–silica core–shell nanoparticles in a dilute aqueous suspension. In this condition the protein itself acts as supramolecular directing agent for silica organization.
To demonstrate this strategy for silica wrapping, we selected β-casein as a model protein. Native β-casein (∼24 kDa, pI ∼ 5.3) is an amphiphilic protein, comprised of a hydrophobic C-terminal domain and a polar, hydrophilic N-terminal domain.44,45 In aqueous solution at room temperature, β-casein self-assembles into micelles with hydrodynamic radii ranging from 12 to 16.3 nm.44–46 These micelles consist of a hydrophobic, dense core and a hydrophilic outer shell of much lower density, quite similar in structure to P123 triblock copolymer micelles used in COK-12 synthesis.43
Experimental
Preparation of β-casein–silica core–shell particles
A typical buffer stock solution with a pH of 5.4 was prepared by dissolving 1.78 g citric acid monohydrate (Sigma-Aldrich) and 12.22 g trisodium citrate dehydrate (Sigma-Aldrich) in 50 g water. 250 mg β-casein from bovine milk (lyophilized powder, Sigma-Aldrich) was dissolved in 50 mL (40 times diluted) citric acid/citrate buffer solution. 1.3 g sodium silicate solution (>27 wt% SiO2, Merck), diluted in 3.75 g demineralized water and 12.5 mL citric acid/citrate buffer stock solution were added to the buffered β-casein solution (Fig. 1). The pH of this suspension increased slightly to approximately 5.9. The final solution was stirred for 5 min and was aged for one day at room temperature without agitation. The first day it remained transparent, but later it became an opaque suspension undergoing coagulation. A powder was obtained by lyophilization. For the 1H NMR DOSY experiment, this method was adjusted by volumetrically replacing H2O with D2O.Characterization
Proton-decoupled 29Si magic angle spinning (MAS) NMR spectra were recorded using a DSX400 spectrometer (Bruker) equipped with a 4 mm MAS probehead. Cross-polarization spectra were obtained using a ramped-shaped contact pulse of 10 ms on the proton channel. For direct-excitation spectra, a single 45° pulse of 2 μs and an interscan delay of 60 s was used and about 6000 scans were accumulated.1H NMR Diffusion-ordered spectroscopy (DOSY) was performed on a 500 MHz spectrometer-Avance III, (Bruker) equipped with a cryo-probe, developing a pulse field gradient of 5 G cm−1 A−1. The sample was thermostated at 300 K. Diffusion NMR data were acquired using a Double Stimulated Echo pulse sequence with bipolar z gradients, for limiting convection effects on self-diffusion measurements. Limited eddy current delay was fixed to 5 ms. The strength and duration of the sinusoidal gradients were optimized for each sample, and varied between 2.3 and 43.3 G cm−1, for a half-gradient delay of 1850 μs. The gradient recovery delay was 200 μs. For each data set, 30k complex points were collected for each of the 40 experiments in which the gradient strength was linearly incremented. A recycling delay of 2 s was respected between scans. DOSY spectra were processed by use of the DOSY module of the software NMRNotebook,47,48 using Inverse Laplace Transform (ILT) driven by maximum entropy, to build the diffusion dimension. An exponential line broadening apodization of 2 Hz was applied to the spectral axis and baseline offset was corrected before DOSY calculation. Intensities of selected NMR peaks were processed by ILT. The final DOSY spectra were obtained with 128 points in the diffusion dimension and 1000 MaxEnt iterations.
Dynamic Light Scattering (DLS) analyses of suspensions were conducted in polystyrene cuvettes at 25 °C on a 90Plus instrument (Brookhaven Instruments Corporation), at a scattering angle of 90° using a 659 nm laser. The fluctuations in the scattered laser light intensity were correlated between 5 μs and 1 s. Correlation functions were analyzed with Igor Pro 6.2, using the Clementine package for modelling decay kinetics based on the Maximum Entropy method. The decay times were converted to hydrodynamic radii with the Stokes–Einstein relation, yielding intensity weighted size distributions.
The β-casein–silica hybrid material was further characterized with specific electron microscopy techniques. Environmental scanning electron microscope images were taken with an XL30 ESEM FEG from FEI (Philips). High angle annular dark field scanning transmission electron microscopy (HAADF-STEM) analysis of β-casein–silica nanocapsules was performed using a Tecnai F20 S/TEM (FEI), operated at 200 kV and equipped with an annular dark-field detector (Fischione).
Small angle X-ray Scattering (SAXS) of native β-casein solution and β-casein–silica nanocapsule suspension were measured at room temperature in a 1 mm quartz capillary on a SAXSess MC2 instrument (Anton Paar) with line-collimated CuKα radiation. Image plate detector and CCD detector were used for β-casein solution and β-casein–silica nanocapsules respectively.
Results and discussion
29Si MAS NMR
The degree of silica condensation in the freeze dried β-casein–silica hybrid material was investigated using 29Si MAS NMR. Cross-polarization (CP) and direct-excitation 29Si MAS NMR spectra are shown in Fig. 2. 1H–29Si cross-polarization (CP) is used to enhance the 29Si NMR signal intensity, and record 29Si NMR spectra in relatively short time. The CP spectrum shows the overlapping signals of different types of silicon atoms Qn = Si(OSi)n(OH)4−n, with strong signals for n = 2 and 3, typical for an incompletely condensed silica network. Quantification of Si-atom types via single-pulse excitation (SPE) 29Si NMR confirmed the abundance of incompletely condensed silica. | ||
| Fig. 2 Cross-polarization (black) and direct-excitation (grey) 29Si MAS NMR spectra of freeze dried β-casein–silica hybrid material. | ||
Diffusion-ordered spectroscopy (DOSY)
To demonstrate the long-lived association between β-casein and the silica in suspension, the protein self-diffusion was probed by use of 1H NMR diffusion-ordered spectroscopy (DOSY). This NMR method, which is a pulsed field gradient (PFG) NMR modulated experiment, is conventionally employed to investigate the individual self-diffusion of molecules and molecular aggregates in mixed solutions down to diffusivities of 0.2 × 10−10 m2 s−1. Our study involves dilute suspensions of 0.4 wt% β-casein. This low concentration is proposed to form separate β-casein–silica nanostructures, rather than embedding the protein or protein aggregates in a continuous silica matrix. The low concentration of these protein suspensions with and without silica poses strong demands on NMR sensitivity. For the highest NMR signal-to-noise ratio in our DOSY experiments a helium-cooled NMR cryo-probe was used, while the sample was kept at room temperature. As a consequence of the electronic design of such NMR cryo-probes, their use for DOSY inevitably introduces some temperature gradients in the sample. To suppress the influence of thermally driven convection on the self-diffusion NMR measurements the Double-Stimulated Gradient Echo (DSGE) pulse sequence proposed by Jerschow and Müller49 was used for collecting the DOSY spectra. 1H NMR spectra of β-casein suspensions with and without silica in citrate-buffered D2O consist of overlapping signals from the protein, the citrate buffer and deuterium exchanged water traces, HDO (Fig. 3A and D). The β-casein-methyl signals at 0.8 ppm have no overlap with other signals, and are, therefore, selected for DOSY analysis as measure for protein mobility. Since the overall β-casein concentration in suspensions with and without silica are virtually equal, the casein-methyl signals are of equal intensity. For both the β-casein with and without silica, the casein-methyl signals decay sufficiently in function of the gradient strength up to 0.43 T m−1, allowing proper inverse Laplace transform analysis. The resulting 2D spectra with chemical shift along the horizontal axis and the self-diffusion coefficient along the vertical axis are shown in Fig. 3. As can be seen in Fig. 3C, the methyl signals of the pure casein suspension are associated with a diffusion coefficient of ca. 1.0 × 10−10 m2 s−1, which may be compared to 6.0 × 10−10 m2 s−1 for the citrate/citric acid molecules. In the suspension with silica, the casein and citrate self-diffusion are 0.9 × 10−10 and 5.5 × 10−10 m2 s−1, respectively (Fig. 3F). These values are both ca. 10% lower than the corresponding components in the β-casein suspension without silica, suggesting a slight increase in viscosity.Self-diffusion of particles or nanostructures depends on viscosity and the particle characteristics, notably its mass and shape.50–52 β-Casein micelles and β-casein–silica hybrid nanostructures can provisionally be approximated as spherical objects for the DOSY analysis.51,52 The similar protein self-diffusion in the β-casein suspensions with and without silica, therefore indicates that the size of the micelles in the pure β-casein solution is comparable to that of the protein-containing nanostructures, possibly associated with silica, in the β-casein–silica system. The casein-methyl signals in the first DOSY trace recorded with the weakest gradient for the casein–silica suspension (Fig. 3E) decreased by approximately 40% compared to the corresponding trace for the pure casein system (Fig. 3B). This indicates part of the β-casein is not observed in DOSY, as result of NMR relaxation during the DSTE pulse sequence. This missing part probably reflects a casein fraction with a shorter relaxation, consistent with casein residing in larger nanostructures, not observable in DOSY. This observation motivated further investigation of the size distribution by DLS.
Dynamic light scattering
DLS was used to determine the hydrodynamic radius distribution in the 0.4 wt% β-casein suspensions with and without sodium silicate. The scattering correlation function, weighed with the R6 proportionality of the scattering per particle, of the pure casein solution corresponds to a log-Gaussian radius distribution centred at 13.8 nm (Fig. 4). This is the typical size for β-casein micelles in dilute solution.44–46 The light scattering after addition of sodium silicate is consistent with a bi-modal distribution of hydrodynamic radii, with a major number-based fraction of smaller structures, with radii distributed around 9.3, and a minor fraction with larger radii around 36 nm. Note that the scattering-intensity distribution in Fig. 4 reflects the hydrodynamic-size distribution weighed by the R6 proportionality. | ||
| Fig. 4 Intensity weighted size distribution (hydrodynamic-radii (RDLS)) of the 0.4 wt% β-casein suspension before and after addition of sodium silicate, monitored by dynamic light scattering. | ||
Electron microscopy
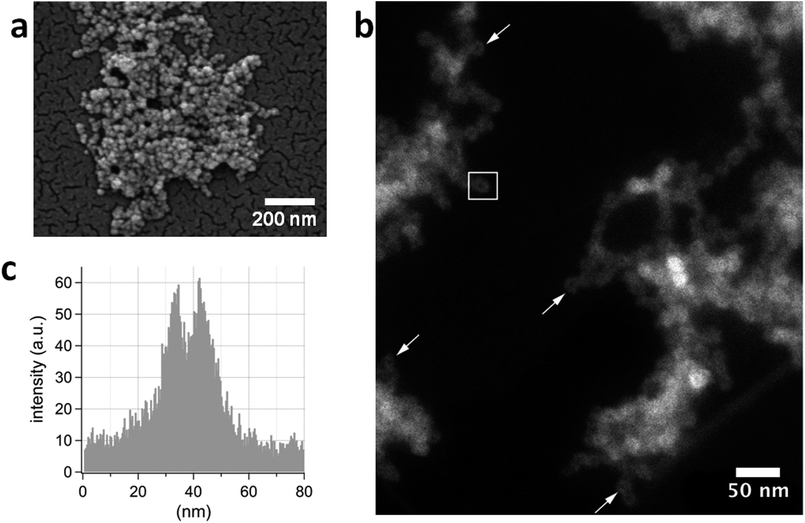
Environmental scanning electron microscopy (ESEM) images of β-casein–silica suspension spread on the sample holder reveal spherical, 15–20 nm large nanoparticles with strictly limited polydispersity (Fig. 5a). Further analysis with high angle annular dark field scanning transmission electron microscopy (HAADF-STEM) shows nanoparticles (Fig. 5b) with an estimated mean particle radius of 8.8 ± 1.2 nm, in agreement with DLS and ESEM measurements.Energy dispersive X-ray analysis of single nanoparticles proved that Si is an abundant constituent element. In HAADF-STEM images, the intensity is approximately proportional to the square of the atomic number Z and also correlates with sample thickness. In Fig. 5b, isolated nanoparticles yield lower intensity at their centre in comparison to their rim. This intensity variation is also observable in Fig. 5c, where an intensity profile, taken across the particle indicated by the white rectangle in Fig. 5b is presented. Similar intensity profiles were obtained for 68 investigated nanoparticles and a mean core radius of 6.6 ± 1.2 nm was determined. The HAADF-STEM observations provide strong evidence that the casein–silicate hybrid nanoparticles are in fact individual core–shell particles, consisting of a dense silica shell surrounding a less dense core.
Small angle X-ray scattering
To further characterize the structure of β-casein micelles and β-casein–silica nanoparticles in the undisturbed suspension, SAXS patterns were recorded (Fig. 6). The SAXS pattern of β-casein micelles without silica can be adequately described by polydisperse spherical particles, comprising a dense core radius of 5.2 nm plus a low density outer shell with a thickness of 7.8 nm, resulting in a total radius of 13.0 nm. The difference in electron density between the protein core and outer shell is small, indicating only a small difference in protein density. These dimensions are in agreement with previously reported radii for β-casein micelles in small angle scattering experiments.44 The particle radius observed by SAXS of approximately 13.0 nm is lower than the hydrodynamic radius derived from DLS, being 13.8 nm. This difference is also reported by others and can be interpreted as the presence of a solvation layer reducing the diffusivity of the micelles or else by the presence of loosely adsorbed protein components on the micelle.53The recorded SAXS intensity of suspended β-casein–silica nanoparticles originates largely from the silicate material, because of its higher contrast to aqueous buffer solution as well as to proteins. At small length scales (large q, >0.02 Å−1), the SAXS pattern is satisfactorily described with a model of polydisperse spherical core–shell particles (Fig. 6). Note that the contrast difference used in the modelling for the free β-casein micelles, originates from the electron density difference between the solvent, the hydrophobic protein core and a less dense outer protein layer. In the latter case, contrast in the core–shell model derives from electron densities correlated with solvent, protein and silica. From the model fit of the casein–silica hybrid in suspension, an average core radius of 7 nm has been determined. This value is in close agreement with an inner radius of 6.6 nm derived from the HAADF-STEM intensity profiles. At q = 0.5 Å−1 a distinct feature in the pattern is observed. This is reflected in a limited size polydispersity of 0.24 in the model fit. The core–shell particles have an average shell thickness of 1.5 nm. Adding this value to the core radius leads to a total average particle radius of 8.5 nm.
At small q-values (q < 0.02 Å−1), information regarding length scales larger than the individual β-casein core–silica shell particles are obtained. The SAXS intensity decays according to a power law with an exponent of −1.8, indicating that the casein–silica particles in suspension had formed open, fractal aggregates by diffusion-limited aggregation.
Synopsis
Combined, the results from the characterization techniques confirm the formation of separate β-casein core–silica shell particles. 29Si MAS NMR of the lyophilized β-casein–silica suspensions show that the silicate molecules actually condense into silicate polymer structures at the low concentration and the employed mild temperature and pH conditions. 1H NMR DOSY reveals protein self-diffusion in the 0.4 wt% β-casein suspension of 1.0 × 10−10 m2 s−1. This diffusivity deviates from reported, much slower micelle self-diffusion in 8 and 15 wt% native-casein gels, respectively 3.4 × 10−12 and 2 × 10−13 m2 s−1.54,55 However, such micelles, which are composed of mixed casein types (including αs1-, αs2-, β-, κ-casein), have a large range in sizes up to a radius 150 nm. The faster self-diffusion observed for the pure β-casein micelles with their hydrodynamic radius of 13.4 nm according to DLS is actually quite consistent, especially if the lower viscosity of the 0.4 wt% suspension is also taken into account.Intuitively, one may have expected that association of β-casein and silica should slow down protein mobility. Self-diffusion of particles or nanostructures in a continuous medium is controlled by viscosity, particle geometry, and its mass. Similar protein self-diffusion in the casein suspensions with and without silica, clearly indicates that the size of the micelles in the pure casein suspension is comparable to that of the protein-containing nanostructures in the casein–silica system.
Careful comparison of DOSY with quantitative 1D 1H NMR spectra reveals that a certain casein fraction is not observable in DOSY, due to a modification of the relaxation for a fraction of the sites. This distribution of small nanostructures, similar in size to the original β-casein micelles, and larger nanostructures, is confirmed by DLS and SAXS. Whereas 29Si MAS NMR, 1H DOSY and DLS do not give unambiguous evidence for the organization of β-casein and silica, electron microscopy and small-angle X-ray scattering reveal the presence of core–shell particles with a dense shell and a diameter comparable to the initial β-casein micelles. Part of these are aggregated, which explains the failure to detect the whole protein content by NMR. The decrease in the radius of individual particles from 13 to 8.5 nm observed by SAXS upon addition of silica to β-casein micelles can be explained, assuming the low density outer layer of the β-casein micelles is pervious to silica. In this case, silica shell formation occurs at the interface of the hydrophobic β-casein core and its hydrophilic outer layer, leaving at least part of the low density protein shell intact and embedded in the silica shell. This is also in agreement with the decrease in particle radius from 13.8 to 9.3 nm upon silica wrapping as determined from DLS. The agreement between the β-casein–silica particle sizes extracted from SAXS and DLS suggests that little or no adsorbed protein molecules are attached to the external surface of the nanocapsules. Fig. 7 gives an overview of all sizes for the encapsulation of β-casein measured with SAXS, TEM and DLS before and after reaction with sodium silicate.
As a consistency check of the core–shell model with the parameters indicated in Fig. 7, a rough estimate of the protein density in the core can be made by also including the overall β-casein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) silica ratio of ∼0.7 (w/w) at the start of the synthesis (0.25 mg β-casein: 1.3 g Na2SiO4 solution containing the equivalent of 27 wt% SiO2; see Experimental section). For a 8.8 nm outer radius of the particles and shell thickness of 1.8 nm the core
silica ratio of ∼0.7 (w/w) at the start of the synthesis (0.25 mg β-casein: 1.3 g Na2SiO4 solution containing the equivalent of 27 wt% SiO2; see Experimental section). For a 8.8 nm outer radius of the particles and shell thickness of 1.8 nm the core![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) shell volume ratio would be ∼1 (v/v). Combination of the gravimetric and volumetric ratio suggests that the density ratio of casein in the core and silica in the shell is roughly 0.7. This density ratio may be compared with the density value of fused silica 2.2 g cm−3 and the typical 1.3 g cm−3 density of a single globular protein.55 The latter represents an upper bound for the protein density in the core. In native casein micelles the protein density actually lies between 0.4 and 0.9 g cm−3.47 The argument above suggests that the protein molecules are closely packed with their hydrophobic parts in the core and their hydrophilic regions partly embedded within the silica shells. The presence of protein tails in the shell, and the incomplete silica condensation could lower the average silica density. However, an explanation may also be that a significant casein fraction is not encapsulated, not al the silica is quantitatively incorporated in the particles or that the silica shells are not forming a completely closed sphere. After all, the picture that the β-casein–silica system forms mono-disperse and perfectly structured core–shell particles is likely to be an oversimplification.
shell volume ratio would be ∼1 (v/v). Combination of the gravimetric and volumetric ratio suggests that the density ratio of casein in the core and silica in the shell is roughly 0.7. This density ratio may be compared with the density value of fused silica 2.2 g cm−3 and the typical 1.3 g cm−3 density of a single globular protein.55 The latter represents an upper bound for the protein density in the core. In native casein micelles the protein density actually lies between 0.4 and 0.9 g cm−3.47 The argument above suggests that the protein molecules are closely packed with their hydrophobic parts in the core and their hydrophilic regions partly embedded within the silica shells. The presence of protein tails in the shell, and the incomplete silica condensation could lower the average silica density. However, an explanation may also be that a significant casein fraction is not encapsulated, not al the silica is quantitatively incorporated in the particles or that the silica shells are not forming a completely closed sphere. After all, the picture that the β-casein–silica system forms mono-disperse and perfectly structured core–shell particles is likely to be an oversimplification.
Conclusions
In conclusion, we propose a single-step method to synthesize protein–silica nanocapsules in dilute aqueous solutions or suspensions using cost-effective, biocompatible sodium silicate and citric acid/citrate buffer. Evidence from DLS, NMR, TEM and SAXS confirms formation of silica shells of a few nanometers thick around discrete self-assembled β-casein micelles, similar to the early stages of the self-assembly process of COK-12 ordered mesoporous silica material using the same conditions and silica source in combination with triblock copolymer.The mild temperature and pH conditions used during encapsulation and the fact that no protein denaturing alcohols are released during the silica formation, hallmark our approach as a biocompatible process. This approach can potentially be extended to a variety of proteins and allows the synthesis of novel silica materials with tailor-made structures and adjustable properties. The straightforward fabrication of these materials broaden the scope of sol–gel science, extending the application field of encapsulation of fragile bio-molecules. Eventually this may offer manifold potential applications in biosensing, biocatalysis or controlled release of therapeutic proteins.
Acknowledgements
JAM and GVT acknowledge the Flemish Government for long-term structural funding (Methusalem). This work was supported by an interuniversity collaboration program (IAP) FT acknowledges KU Leuven for a BOF SF fellowship. The work was supported by the Flemish FWO. RM and AA acknowledge the Flemish FWO for a postdoctoral fellowship. The authors acknowledge financial support from the European Union under the Framework 7 program for the grant for the Integrated Infrastructure Initiative N. 262348 European Soft Matter Infrastructure (ESMI). The authors acknowledge support from the European Research Council (ERC Grant #335078-COLOURATOMS). We thank Heiner Santner of Anton Paar GmbH (Austria) for the SAXS measurements with the SAXSess instrument. We also thank Klaus Zick and Rainer Kesserbaum of Bruker Biospin GmbH (Germany) for the DOSY experiments.References
- J. Livage, T. Coradin and C. Roux, J. Phys. Condens. Matter., 2001, 13, R673–R691 CrossRef CAS.
- A. C. Pierre, Biocatal. Biotransform., 2004, 22, 145–170 CrossRef CAS.
- D. Avnir, T. Coradin, O. Lev and J. Livage, J. Mater. Chem., 2006, 16, 1013–1030 RSC.
- T. R. Besanger and J. D. Brennan, J. Sol-Gel Sci. Technol., 2006, 40, 209–225 CrossRef CAS.
- L. Betancor and H. R. Luckarift, Trends Biotechnol., 2008, 26, 566–572 CrossRef CAS PubMed.
- B. Menaa, F. Menaa, C. Aiolfi-Guimaraes and O. Sharts, Int. J. Nanotechnol., 2010, 7, 1–45 CrossRef CAS.
- Z. Cai, Z. Ye, X. Yang, Y. Chang, H. Wang, Y. Liu and A. Cao, Nanoscale, 2011, 3, 1974–1976 RSC.
- A. Cao, Z. Ye, Z. Cai, E. Dong, X. Yang, G. Liu, X. Deng, Y. Wang, S.-T. Yang, H. Wang, M. Wu and Y. Liu, Angew. Chem., Int. Ed., 2010, 49, 3022–3025 CrossRef CAS PubMed.
- Z. Y. Zhao, J. Liu, M. Hahn, S. Qiao, A. P. J. Middelberg and L. He, RSC Adv., 2013, 3, 22008–22013 RSC.
- I. Hegedus and E. Nagy, Chem. Eng. Sci., 2009, 64, 1053–1060 CrossRef CAS PubMed.
- B. Campanini, S. Bologna, F. Cannone, G. Chirico, A. Mozzarelli and S. Bettati, Protein Sci., 2005, 14, 1125–1133 CrossRef CAS PubMed.
- Z. P. Yang, S. H. Si and C. J. Zhang, Biochem. Biophys. Res. Commun., 2008, 367, 169–175 CrossRef CAS PubMed.
- J. Kim, J. W. Grate and P. Wang, Chem. Eng. Sci., 2006, 61, 1017–1026 CrossRef CAS PubMed.
- J. Kim and J. W. Grate, Nano Lett., 2003, 3, 1219–1222 CrossRef CAS.
- D. Ma, M. Li, A. J. Patil and S. Mann, Adv. Mater., 2004, 16, 1838–1841 CrossRef CAS.
- R. Kumar, A. N. Maitra, P. K. Patanjali and P. Sharma, Biomaterials, 2005, 26, 6743–6753 CrossRef CAS PubMed.
- M. Yan, J. Ge, Z. Liu and P. Ouyang, J. Am. Chem. Soc., 2006, 128, 11008–11009 CrossRef CAS PubMed.
- M. Fujiwara, K. Shiokawa, K. Hayashi, K. Morigaki and Y. Nakahara, J. Biomed. Mater. Res., Part A, 2007, 81, 103–112 CrossRef PubMed.
- B. Hu, M. Li, S. Sadasivan, A. J. Patil and S. Mann, Nanoscale, 2011, 3, 1031–1036 RSC.
- A. J. Patil, M. Li and S. Mann, Nanoscale, 2013, 5, 7161–7174 RSC.
- D. Fujita, K. Suzuki, S. Sato, M. Yagi-Utsumi, Y. Yamaguchi, N. Mizuno, T. Kumasaka, M. Takata, M. Noda, S. Uchiyama, K. Kato and M. Fujita, Nat. Commun., 2012, 3, 1093 CrossRef PubMed.
- M. Yan, J. J. Du, Z. Gu, M. Liang, Y. F. Hu, W. J. Zhang, S. Priceman, L. L. Wu, Z. H. Zhou, Z. Liu, T. Segura, Y. Tang and Y. F. Lu, Nat. Nanotechnol., 2010, 5, 48–53 CrossRef CAS PubMed.
- S. Mann, Nat. Mater., 2009, 8, 781–792 CrossRef CAS PubMed.
- N. E. Botterhuis, Q. Sun, P. C. M. M. Magusin, R. A. van Santen and N. A. J. M. Sommerdijk, Chem.–Eur. J., 2006, 12, 1448–1456 CrossRef CAS PubMed.
- M. Perullini, M. a. Jobbágy, M. Bermúdez Moretti, S. Correa García and S. A. Bilmes, Chem. Mater., 2008, 20, 3015–3021 CrossRef CAS.
- L. Ellerby, C. Nishida, F. Nishida, S. Yamanaka, B. Dunn, J. Valentine and J. Zink, Science, 1992, 255, 1113–1115 CAS.
- J. Livage, C. Roux, J. M. Costa, J. F. Quinson and I. Desportes, J. Sol-Gel Sci. Technol., 1996, 7, 45–51 CrossRef CAS.
- R. Campostrini, G. Carturan, R. Caniato, A. Piovan, R. Filippini, G. Innocenti and E. M. Cappelletti, J. Sol-Gel Sci. Technol., 1996, 7, 87–97 CrossRef CAS.
- M. L. Ferrer, L. Yuste, F. Rojo and F. del Monte, Chem. Mater., 2003, 15, 3614–3618 CrossRef CAS.
- C. Guan, G. Wang, J. Ji, J. Wang, H. Wang and M. Tan, J. Sol-Gel Sci. Technol., 2008, 48, 369–377 CrossRef CAS PubMed.
- S. H. Yang, K.-B. Lee, B. Kong, J.-H. Kim, H.-S. Kim and I. S. Choi, Angew. Chem., Int. Ed., 2009, 48, 9160–9163 CrossRef CAS PubMed.
- G. Wang, L. Wang, P. Liu, Y. Yan, X. Xu
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) and R. Tang
and R. Tang![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) , ChemBioChem, 2010, 11, 2368–2373 CrossRef CAS PubMed.
, ChemBioChem, 2010, 11, 2368–2373 CrossRef CAS PubMed. - N. M. Eleftheriou, X. Ge, J. Kolesnik, S. B. Falconer, R. J. Harris, C. Khursigara, E. D. Brown and J. D. Brennan, Chem. Mater., 2013, 25, 4798–4805 CrossRef CAS.
- J. D. Brennan, Acc. Chem. Res., 2007, 40, 827–835 CrossRef CAS PubMed.
- I. Ichinose, Y. Hashimoto and T. Kunitake, Chem. Lett., 2004, 33, 656–657 CrossRef CAS.
- A. J. Patil, E. Muthusamy and S. Mann, Angew. Chem., Int. Ed., 2004, 43, 4928–4933 CrossRef CAS PubMed.
- D. M. Liu and I. W. Chen, Acta Mater., 1999, 47, 4535–4544 CrossRef CAS.
- R. B. Bhatia, C. J. Brinker, A. K. Gupta and A. K. Singh, Chem. Mater., 2000, 12, 2434–2441 CrossRef CAS.
- T. Coradin, A. Coupe and J. Livage, Colloids Surf., B, 2003, 29, 189–196 CrossRef CAS.
- J. Jammaer, A. Aerts, J. D'Haen, S. Jin Won and J. A. Martens, J. Mater. Chem., 2009, 8290–8293 RSC.
- J. Jammaer, T. S. van Erp, A. Aerts, C. E. A. Kirschhock and J. A. Martens, J. Am. Chem. Soc., 2011, 133, 13737–13745 CrossRef CAS PubMed.
- J. L. Bass and G. L. Turner, J. Phys. Chem. B, 1997, 101, 10638–10644 CrossRef CAS.
- I. Portnaya, R. Khalfin, E. Kesselman, O. Ramon, U. Cogan and D. Danino, Phys. Chem. Chem. Phys., 2011, 13, 3153–3160 RSC.
- E. Leclerc and P. Calmettes, Phys. B, 1997, 241–243, 1141–1143 CrossRef CAS.
- A. Thurn, W. Burchard and R. Niki, Colloid Polym. Sci., 1987, 265, 653–666 CAS.
- J. E. O'Connell, V. Y. Grinberg and C. G. de Kruif, J. Colloid Interface Sci., 2003, 258, 33–39 CrossRef.
- D. Tramesel, V. Catherinot and M.-A. Delsuc, J. Magn. Reson., 2007, 188, 56–67 CrossRef CAS PubMed.
- NMRNotebook, from NMRTEC, http://www.nmrtec.com/software/nmrnotebook.
- A. Jerschow and N. Müller, J. Magn. Reson., 1997, 125, 372–375 CrossRef CAS.
- W. B. Russel, Annu. Rev. Fluid Mech., 1981, 13, 425–455 CrossRef.
- S. Augé, P.-O. Schmit, C. A. Crutchfield, M. T. Islam, D. J. Harris, E. Durand, M. Clemancey, A.-A. Quoineaud, J.-M. Lancelin, Y. Prigent, F. Taulelle and M.-A. Delsuc, J. Phys. Chem. B, 2009, 113, 1914–1918 CrossRef PubMed.
- S. Floquet, S. Brun, J.-F. Lemonnier, M. Henry, M.-A. Delsuc, Y. Prigent, E. Cadot and F. Taulelle, J. Am. Chem. Soc., 2009, 131, 17254–17259 CrossRef CAS PubMed.
- K. Kajiwara, R. Niki, H. Urakawa, Y. Hiragi, N. Donkai and M. Nagura, Biochim. Biophys. Acta, 1988, 955, 128–134 CrossRef CAS.
- S. Le Feunteun, M. Ouethrani and F. Mariette, Food Hydrocolloids, 2012, 27, 456–463 CrossRef CAS PubMed.
- F. Mariette, D. Topgaard, B. Jönsson and O. Soderman, J. Agric. Food Chem., 2002, 50, 4295–4302 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2014 |