Bilayer interaction and protein kinase C-C1 domain binding studies of kojic acid esters†
Abstract
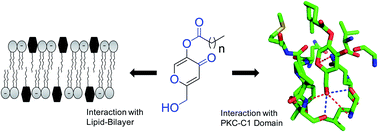
Development of protein kinase C (PKC) regulators has been considered as an attractive therapeutic strategy for the treatment of cancer and other diseases. Extensive efforts are underway to synthesize PKC regulators targeted to the DAG-responsive C1 domain. Investigation of physicochemical properties of the synthesized molecules also is essential for the development of PKC-C1 domain ligands. To develop PKC regulators, we conveniently synthesized kojic acid esters targeted to the DAG/phorbol ester binding site within the C1 domain. Physicochemical studies showed that the kojic acid esters aggregate in aqueous solution at reasonably lower concentration. The results also showed that the compounds strongly interact with the lipid bilayer and the hydrophilic part of the compound localize at the bilayer/water interface. In vitro protein binding studies and molecular docking analysis revealed that the hydroxymethyl group, carbonyl groups and acyl chain length are important for their interaction with the C1 domain. The potent compound showed more than 10-fold stronger binding affinity for the C1 domain than DAG. In addition to the diverse application of kojic acid esters in food, cosmetic and skin-health industries, these findings reveal that ester analogues represent an attractive group of C1-domain ligands that can be further structurally modified to improve their binding and activity.

 Please wait while we load your content...
Please wait while we load your content...