Facile preparation of graphene/Fe3O4/TiO2 multifunctional composite for highly selective and sensitive enrichment of phosphopeptides†
Yulu Lianga,
Xiwen Hea,
Langxing Chen*a and
Yukui Zhangab
aState Key Laboratory of Medical Chemical Biology, Research Center for Analytical Science, College of Chemistry, Nankai University, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300071, P. R. China. E-mail: lxchen@nankai.edu.cn; Fax: +86-0-22-23502458; Tel: +86-0-22-23505091
bDalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116011, P. R. China. E-mail: ykzhang@dicp.ac.cn; Fax: +86-411-84379560; Tel: +86-411-84379560
First published on 9th April 2014
Abstract
A novel multifunctional graphene/Fe3O4/TiO2 composite with excellent hydrophilicity and biological compatibility was synthesized and applied to the fast, highly selective and sensitive enrichment of phosphopeptides from biosamples.
Protein phosphorylation is one of the most significant post-translational modifications (PTMs) in nature, playing a crucial role in eukaryotic cells, involved in many regulatory functions, such as cell growth, differentiation, division, signal transduction and metabolism.1 Therefore, a comprehensive analysis of protein phosphorylation via the identification of phosphorylation sites is of keen interest in the field of proteomics.2 In recent years, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and electrospray-ionization mass spectrometry (ESI-MS),3 have been widely employed to study PTMs owing to their high sensitivity and high-throughput. However, it is still a great challenge to detect phosphopeptides due to their low abundance and low ionization efficiency.4 Therefore, the selective enrichment of phosphoproteins or phosphopeptides from complex biosamples is an essential step prior to MS analysis.
To date, immobilized metal ion affinity chromatography (IMAC) has been widely applied to enrich the phosphopeptides. When loaded on IMAC, the chelating metal ions (Fe3+, Ga3+, Zr4+, Ti4+) bind specifically to the phosphopeptides from complex mixtures.5 Nevertheless, the limitation in IMAC-based enrichment is the high level of unspecific binding of acidic peptides.6 Metal oxide affinity chromatography (MOAC) has emerged to be an attractive alternative to IMAC.7 Although TiO2-based MOAC materials have been the most commonly used for the enrichment of phosphopeptides, owing to its high chemical stability over a wide pH range, selectivity and recovery, and relatively high salt tolerance,8 the aggregation of TiO2 particles limits the available surface area of adsorbent for the efficient enrichment.
Graphene, an sp2-bonded carbon sheet with the thickness of a single atom, has attracted a great deal of attention in recent years due to its unique mechanical, thermal, optical and electrical properties.9 With the inherent superiorities, graphene has been successfully utilized as a good adsorbent in sample pretreatment based on its ultrahigh specific surface area, high loading capacity, large delocalized π-electron system and hydrophobic interaction.10 However, its poor hydrophilicity and difficulty in the surface functionalization often limit its more extensive bioapplication. Recently, Ti4+ or TiO2 immobilized graphene can be used as the adsorbent for the selective extraction of phosphopeptides from complex peptides mixtures.11 The direct use of graphene as an adsorbent needs a complicated process to isolate the graphene and targets from the samples. Decorating magnetic iron oxide nanoparticles onto graphene in a composite will facilitate to impart the desirable magnetic property into graphene in a variety of application fields, such as bioseparation, medical diagnosis, magnetically targeted drug delivery, magnetic energy storage and catalysis.12
Herein, we designed a facile synthetic strategy of graphene/Fe3O4/TiO2 multifunctional composite material for the enrichment of phosphopeptides (Scheme 1). The graphene/Fe3O4/TiO2 multifunctional composite exhibited several attractive advantages. Firstly, the large specific surface area of graphene offers higher capacity for loading the TiO2 nanoparticles and thus possesses the high available surface area for the efficient enrichment of phosphopeptides. Second, the hydrophilic polydopamine layer on the magnetic graphene exhibits excellent environmental stability, good biocompatibilty and good water dispersibility,13 and also decreases the non-specific adsorption of non-phosphopeptides. Finally, the highly loaded Fe3O4 nanoparticles accelerate the isolation. The resulting graphene/Fe3O4/TiO2 composite can selectively capture, fast magnetically isolate and sequentially determine of the low-abundance phosphopeptides from the complex biosamples.
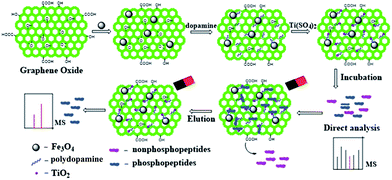 | ||
| Scheme 1 Schematic illustration of the synthesis and enrichment of graphene/Fe3O4/TiO2 multifunctional composite for phosphopeptides. | ||
The synthetic strategy of graphene/Fe3O4/TiO2 magnetic multifunctional composite and the procedure for the selective enrichment of phosphopeptides are illustrated in Scheme 1. The graphene/Fe3O4 composite was firstly prepared by a modified solvothermal method. The surface modified graphene oxide (GO) highly loaded with Fe3O4 nanoparticles is hydrothermally produced from the reduction reaction between FeCl3 and ethylene glycol in the presence of GO. Then, the polydopamine (PDA)-capped graphene/Fe3O4 was prepared via the self-polymerization of dopamine hydrochloride. Dopamine, commonly known as a neuroendocrine transmitter and a unique molecule mimicking the adhesive proteins, exhibits excellent affinity for most organic and inorganic surfaces such as metal, metal oxide, and polymer surfaces and has been found to be able to polymerize into a unique hydrophilic PDA coating on a variety of substrates at a weak alkaline pH.14 Moreover, as a good reducing agent, dopamine has recently been used to fabricate nanocomposites via directly reacting with HAuCl4.15 Finally, graphene/Fe3O4/TiO2 multifunctional composites were obtained by the hydrolysis of Ti4+ immobilized on the PDA (see Experimental details in ESI†).
The TEM images of graphene/Fe3O4 and graphene/Fe3O4/TiO2 composites are shown in Fig. 1. Obviously, the two-dimensional graphene sheets are well decorated by a large quantity of uniform spherical and evenly distributed Fe3O4 particles (with average size of about 150 nm). The TEM image of graphene/Fe3O4/TiO2 (Fig. 1b) clearly reveals that the TiO2 nanoparticles with average diameter of 2 nm highly loaded on the surface of PDA–graphene/Fe3O4. It is clear that the plane structure of graphene/Fe3O4 was well retained due to the good chemical and physical stability of graphene, and the functional layer of PDA was successfully coated on graphene and Fe3O4.
The successful synthesis of graphene/Fe3O4/TiO2 can be further confirmed by Fourier transform infrared spectroscopy (FT-IR), X-ray photoelectron spectroscopy (XPS), and power X-ray diffraction (XRD) analysis. The band at 1083 cm−1 and 877.7 cm−1 was attributed to C–O stretching of the epoxy structure in GO, while the bands at 1618.2 cm−1 and 1187.5 cm−1 can be attributed to the stretching vibrations of C–O in asymmetric and symmetric COO− on the graphene/Fe3O4 (Fig. S1, ESI†). After coating with polydopamine, the bands at 1083 and 877.7 cm−1 disappear, and many new peaks can be seen in PDA–graphene/Fe3O4 (Fig. S1b, ESI†). The peak at 1203 cm−1 was assigned to the C–O stretching of phenolic groups in polydopamine, which indicated that graphene/Fe3O4 had been successfully modified by PDA coating. It was obvious that C, O, Fe, Si, N and Ti all appeared on the surface of graphene/Fe3O4/TiO2 composite from XPS patterns (Fig. S2a, ESI†). A C1s peak around 291.7 eV, O1s peak at 530.6 eV, N1s peak at 398.5 eV, Fe2p peak at 710.2 eV and Ti2p peaks at 459.6 and 453.9 eV can be observed. By the deconvolution of C1s spectrum, three peaks at 284.59, 286.39 and 288.59 eV were identified, which are assigned to the C–C or C–H, C–N or O–C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and benzene rings respectively (Fig. S2b, ESI†). The XRD results show that the diffraction peaks of graphene/Fe3O4/TiO2 matched well with those of magnetite (JCPDS no. 75-1610) (Fig. S3, ESI†). This suggests the synthetic process did not change the crystalline phase of magnetite. The magnetic properties of graphene/Fe3O4/TiO2 composite were studied using a vibrating sample magnetometer (VSM). The magnetic hysteresis loops show that graphene/Fe3O4/TiO2 composite has a fairly strong magnetization (Fig. S4, ESI†) and can be rapidly separated from the mixture within 1 min in the presence of an external magnet (Fig. S4 inset, ESI†).
C, and benzene rings respectively (Fig. S2b, ESI†). The XRD results show that the diffraction peaks of graphene/Fe3O4/TiO2 matched well with those of magnetite (JCPDS no. 75-1610) (Fig. S3, ESI†). This suggests the synthetic process did not change the crystalline phase of magnetite. The magnetic properties of graphene/Fe3O4/TiO2 composite were studied using a vibrating sample magnetometer (VSM). The magnetic hysteresis loops show that graphene/Fe3O4/TiO2 composite has a fairly strong magnetization (Fig. S4, ESI†) and can be rapidly separated from the mixture within 1 min in the presence of an external magnet (Fig. S4 inset, ESI†).
The enrichment performance of graphene/Fe3O4/TiO2 composite for phosphopeptides was first examined by the tryptic digest product of β-casein. From the direct analysis of the digest product (20 fmol), it can be seen that only three weak signals of them originating from phosphopeptides could be detected, while the peaks corresponding to nonphosphorylated peptides dominate (Fig. 2a). After the enrichment using graphene/Fe3O4/TiO2 composite, the signals of those at m/z 1981.8, 2061.8, 2432.0, 2556.8, 3042.2 and 3122.1 which represent phosphopeptide residues derived from β-casein16 can be found (Fig. 2b). Table S1 (ESI†) lists the detailed amino acid sequences of these identified phosphopeptides. When the amount of the digest product decrease to 20 amol and 2 amol, the phosphopeptide peaks at m/z 1981.8, 2061.8, 2556.8 and 3122.1 could still be observed in the mass spectrum after the enrichment using graphene/Fe3O4/TiO2 composite (Fig. 2c and d). As the amount of digest product decreases to 0.2 amol, it is difficult to distinguish the peaks because of the background (Fig. S5, ESI†). The results indicate that graphene/Fe3O4/TiO2 multifunctional composite can specifically extract and completely elute phosphopeptides from the digest solution with high sensitivity.
The capacity of graphene/Fe3O4/TiO2 composite to selectively trap phosphopeptides was further investigated by analyzing the mixture of tryptic digests of β-casein (20 fmol) and BSA at molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. As shown in Fig. S6a (ESI†), nearly no phosphopeptide signal can be observed in the β-casein/BSA tryptic digest mixture at molar ratio of 1
500. As shown in Fig. S6a (ESI†), nearly no phosphopeptide signal can be observed in the β-casein/BSA tryptic digest mixture at molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. Because the introduction of BSA dramatically increases the complexity of the sample, the signals of phosphopeptides are severely suppressed by the greater amount of nonphosphopeptides largely originating from BSA. However after the enrichment with graphene/Fe3O4/TiO2 composite, five intensive peaks corresponding to specific phosphopeptides of β-casein are observed. When adjusting β-casein/BSA to 1
100. Because the introduction of BSA dramatically increases the complexity of the sample, the signals of phosphopeptides are severely suppressed by the greater amount of nonphosphopeptides largely originating from BSA. However after the enrichment with graphene/Fe3O4/TiO2 composite, five intensive peaks corresponding to specific phosphopeptides of β-casein are observed. When adjusting β-casein/BSA to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 with the concentration of β-casein kept at 20 fmol, after the enrichment, the mass spectra revealed that three signals of phosphopeptides of m/z at 2061.8, 2556.1 and 3122.3 (Fig. S6c, ESI†) can be still observed under the clear background. This proves that graphene/Fe3O4/TiO2 composite possessed the excellent capability for the selective enrichment of phosphopeptides from a complex in the presence of large excess of nonphosphopeptide.
500 with the concentration of β-casein kept at 20 fmol, after the enrichment, the mass spectra revealed that three signals of phosphopeptides of m/z at 2061.8, 2556.1 and 3122.3 (Fig. S6c, ESI†) can be still observed under the clear background. This proves that graphene/Fe3O4/TiO2 composite possessed the excellent capability for the selective enrichment of phosphopeptides from a complex in the presence of large excess of nonphosphopeptide.
It is demonstrated that the graphene/Fe3O4/TiO2 composite can be applied in the enrichment of phosphopeptides from the tryptic digest of nonfat milk commonly containing α-casein and β-casein. As shown in Fig. 3a, before the enrichment, only a few signals with low intensity from phosphopeptides were identified, which are almost submerged by those of nonphosphopeptides. However, after the enrichment using graphene/Fe3O4/TiO2 composite, eleven phosphopeptides peaks were identified (seven from α-casein and four from β-casein) with good resolution8a (Fig. 3b) and the corresponding sequences were listed in Table S2 (ESI†). This result demonstrated that graphene/Fe3O4/TiO2 composite can be successfully used for the selective enrichment of phosphopeptides from a very complex real sample.
In summary, a new type of graphene/Fe3O4/TiO2 composite with excellent hydrophilicity and biological compatibility has been synthesized. The multifunctional material was successfully used to highly selectively and sensitively enrich phosphopeptides. Thus, there is promise that the newly synthesized graphene/Fe3O4/TiO2 composite enables a more efficient enrichment of phosphopeptides from a complex peptide mixture in phosphoproteome research.
Acknowledgements
We gratefully appreciate the financial support by the National Basic Research Program of China (no. 2012CB910601), the National Natural Science Foundation of China (no. 20935001, 21275080) and the Research Fund for the Doctoral Program of Higher Education of China (no. 20120031110007).Notes and references
- (a) T. Hunter, Cell, 2000, 100, 113 CrossRef CAS; (b) B. Kersten, G. K. Agrawal and H. Iwahashi, Proteomics, 2006, 6, 5517 CrossRef CAS PubMed; (c) C. L. Nilsson, Anal. Chem., 2012, 84, 735 CrossRef CAS PubMed.
- (a) J. Ptacek, G. Devgan and G. Michaud, Nature, 2005, 438, 679 CrossRef CAS PubMed; (b) Y. Li, X. Xu, D. Qi, C. Deng, P. Yang and X. Zhang, J. Proteome Res., 2008, 7, 2526 CrossRef CAS PubMed.
- (a) J. Tang, P. Yin, X. Lu, D. Qi, Y. Mao, C. Deng, P. Yang and X. Zhang, J. Chromatogr. A, 2010, 1217, 2197 CrossRef CAS PubMed; (b) Z. Lu, L. He and Y. Yin, Chem. Commun., 2010, 46, 6174 RSC.
- G. Han, M. Ye and H. Zou, Analyst, 2008, 133, 1128 RSC.
- (a) T. E. Thingholm, O. N. Jensen and P. J. Robinson, Mol. Cell. Proteomics, 2008, 7, 661 CrossRef CAS PubMed; (b) J. Lu, Y. Li and C. Deng, Nanoscale, 2011, 3, 1225 RSC; (c) X. S. Li, L. D. Xu, G. T. Zhu, B. F. Yuan and Y. Q. Feng, Analyst, 2012, 137, 959 RSC; (d) H. Q. Qin, F. J. Wang, P. Y. Wang, L. Zhao, J. Zhu, Q. H. Yang, R. A. Wu, M. L. Ye and H. F. Zou, Chem. Commun., 2012, 48, 961 RSC; (e) L. Y. Zhang, Q. Zhao, Z. Liang, K. G. Yang, L. L. Sun, L. H. Zhang and Y. K. Zhang, Chem. Commun., 2012, 48, 6274 RSC.
- A. Leitner, M. Sturm and W. Lindner, Anal. Chim. Acta, 2011, 703, 19 CrossRef CAS PubMed.
- (a) L. P. Li, T. Zheng, L. N. Xu, Z. Li, L. D. Sun, Z. X. Nie, Y. Bai and H. W. Liu, Chem. Commun., 2013, 49, 1762 RSC; (b) Y. J. Tan, D. X. Sui, W. H. Wang, M. H. Kuo, G. E. Reid and M. L. Bruening, Anal. Chem., 2013, 85, 5699 CrossRef CAS PubMed; (c) Y. A. Leitner, TrAC, Trends Anal. Chem., 2010, 29, 177 CrossRef PubMed; (d) M. R. Larsen, T. E. Thingholm, O. N. Jensen, P. Roepstorff and T. J. D. Jorgensen, Mol. Cell. Proteomics, 2005, 4, 873 CrossRef CAS PubMed.
- (a) S. T. Wang, M. Y. Wang, X. Su, B. F. Yuan and Y. Q. Feng, Anal. Chem., 2012, 84, 7763 CrossRef CAS PubMed; (b) Y. Y. Zeng, H. J. Chen, K. J. Shiau, S. U. Hung, Y. S. Wang and C. C. Wu, Proteomics, 2012, 12, 280 CrossRef PubMed.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183 CrossRef CAS PubMed.
- (a) Q. Liu, J. B. Shi, J. T. Sun, T. Wang, L. X. Zeng and G. B. Jiang, Angew. Chem., Int. Ed., 2011, 50, 1 CrossRef PubMed; (b) Q. Liu, J. B. Shi and G. B. Jiang, TrAC, Trends Anal. Chem., 2012, 37, 1 CrossRef CAS PubMed.
- (a) L. A. L. Tang, J. Z. Wang, T. K. Lim, X. Z. Bi, W. C. Lee, Q. S. Lin, Y. T. Chang, C. T. Lim and K. P. Loh, Anal. Chem., 2012, 84, 6693 CrossRef CAS PubMed; (b) Y. H. Yan, Z. F. Zheng, C. H. Deng, Y. Li, X. M. Zhang and P. Y. Yang, Anal. Chem., 2013, 85, 8483 CrossRef CAS PubMed; (c) J. Lu, M. Wang, Y. Li and C. H. Deng, Nanoscale, 2012, 4, 1577 RSC.
- (a) C. Y. Shi, J. R. Meng and C. H. Deng, Chem. Commun., 2012, 48, 2418 RSC; (b) G. Cheng, Z. G. Wang, Y. L. Liu, J. L. Zhang, D. H. Sun and J. Z. Ni, Chem. Commun., 2012, 48, 10240 RSC; (c) H. P. Yiu, H. J. Niu and E. Biermans, Adv. Funct. Mater., 2010, 20, 1599 CrossRef CAS; (d) E. Q. Song, J. Hu and C. Y. Wen, ACS Nano, 2011, 5, 761 CrossRef CAS PubMed; (e) N. A. Frey, S. Peng and K. Cheng, Chem. Soc. Rev., 2009, 38, 2532 RSC.
- (a) J. Sedó, J. Saiz-Poseu, F. Busqué and D. Ruiz-Molina, Adv. Mater., 2013, 25, 653 CrossRef PubMed; (b) S. M. Kang, S. Park, D. Kim, S. Y. Park, R. S. Ruoff and H. S. Lee, Adv. Funct. Mater., 2011, 21, 108 CrossRef CAS.
- (a) R. M. Wightman, L. J. May and A. C. Michael, Anal. Chem., 1988, 60, 769 CrossRef; (b) H. Lee, S. M. Dellatore and W. M. Miller, Science, 2007, 318, 426 CrossRef CAS PubMed; (c) S. M. Kang, J. Rho and I. S. Choi, J. Am. Chem. Soc., 2009, 131, 13224 CrossRef CAS PubMed; (d) M. Zhang, X. H. Zhang, X. W. He, L. X. Chen and Y. K. Zhang, Nanoscale, 2012, 4, 3141 RSC.
- (a) L. Q. Guo, Q. Liu, G. L. Li, J. B. Shi, J. Y. Liu, T. Wang and G. B. Jiang, Nanoscale, 2012, 4, 5864 RSC; (b) M. Zhang, X. W. He, L. X. Chen and Y. K. Zhang, J. Mater. Chem., 2010, 20, 10696 RSC.
- A. Nita-Lazar, H. Saito-Benz and F. M. White, Proteomics, 2008, 8, 4433 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The experimental details and additional figures and tables. See DOI: 10.1039/c4ra01573h |
| This journal is © The Royal Society of Chemistry 2014 |



