Hyaluronic acid–siRNA conjugates complexed with cationic solid lipid nanoparticles for target specific gene silencing
Min-Young Lee,
Won Ho Kong,
Ho Sang Jung and
Sei Kwang Hahn*
Department of Materials Science and Engineering, Pohang University of Science and Technology (POSTECH), San 31 Hyoja-dong, Nam-gu, Pohang, Kyungbuk 790-784, Korea. E-mail: skhanb@postech.ac.kr; Fax: +82-54-279-2399; Tel: +82-54-279-2159
First published on 14th April 2014
Abstract
Despite extensive investigations on siRNA delivery systems for the past decade, there has been no clinically available product until now. In this work, reducible hyaluronic acid (HA)–siRNA conjugate was successfully synthesized and used to make a complex with cationic solid lipid nanoparticles (CSLNs) for the development of a liver specific siRNA delivery system. The reducible HA–siRNA conjugate was synthesized by the disulfide–thiol exchange reaction between pyridyldithiol modified HA and thiolated siRNA. The remaining pyridyldithiol was further blocked with cysteine. The biomimetic CSLNs were prepared by reconstituting the composition of natural apolipoprotein-free low density lipoproteins (LDLs). The formation of the HA–siRNA/CSLN complex was confirmed by gel electrophoresis (GE), dynamic light scattering (DLS), and atomic force microscopy (AFM). The HA–siRNA/CSLN complex showed remarkably low cytotoxicity and high transfection efficiency in the presence of serum. The therapeutic index (LC50/IC50) of the HA–siRNA/CSLN complex was statistically much higher than that of a HA–siRNA conjugate or siRNA complexed with commercially available siRNA transfection reagents like in vivo jetPEI and INTERFERin, as well as an siRNA/CSLN complex. The HA–siRNA/CSLN complex can be effectively applied as a model system for the treatment of liver diseases, such as liver fibrosis and liver cancer.
Introduction
Since siRNA was reported in 1999, its delivery system has been widely investigated. Nevertheless, there has been no clinically available product until now.1,2 The development of target specific and safe delivery systems of siRNA is the prerequisite for further clinical applications.3 Although some big pharmaceutical companies have shied away from the development of RNAi therapeutics due to the difficulty in its delivery, it still has huge potential for the treatment of various diseases caused by genetic disorders and viral infection.3,4 The ineffective intracellular delivery of siRNA might be caused by its negative charge, short length, high stiffness, and fast degradation in the serum.3,4 To date, a variety of materials, such as cationic polymers, lipid-like carriers, and nanoparticles, have been used for the formation of a complex with siRNA to overcome the technical hurdles.5–7 In addition, siRNA conjugates, such as cholesterol–siRNA,8,9 α-tocopherol–siRNA,10 lipid–siRNA,11 peptide–siRNA,12,13 aptamer–siRNA,14 and poly(ethylene glycol)–siRNA conjugates,15,16 have been reported to improve their pharmacokinetic behaviours, cellular uptake, and target specificity.17 Furthermore, self-crosslinked and multimerized siRNA has been developed to make more stable and compact polyelectrolyte complexes.3Hyaluronic acid (HA) is a negatively charged natural biopolymer and has several advantages as a drug delivery carrier including the negligible non-specific interaction with serum components and the highly efficient target specific delivery to the liver tissues with HA receptors like the cluster determinant 44 (CD44), hyaluronan receptor for endocytosis (HARE), and so on.4,18 We previously reported that reducible polyethyleneimine (rPEI)–HA conjugate complexed with siRNA showed a feasible therapeutic effect on liver cirrhosis19 and reducible HA–siRNA conjugate complexed with relatively non-toxic linear PEI was target specifically delivered to the liver, resulting in the significant liver-specific gene silencing.20 The conjugation of siRNA with HA contributed to reduce its degradation in the serum and facilitate its target specific delivery to the liver. We also developed cationic solid lipid nanoparticles (CSLNs) derived from apolipoprotein-free low density lipoproteins (LDLs), which can be recognized by lipoprotein receptors in the liver like LDL receptor and remnant receptor, to deliver siRNA specifically into the liver and treat liver fibrosis.21
In this work, based on our previous studies, we tried to develop a reducible HA–siRNA conjugate/CSLN complex with low cytotoxicity and high transfection efficiency for the liver-specific gene silencing by the dual targeting effect of HA and CSLN. As reported in our previous article,18 the reducible HA–siRNA conjugate was synthesized by the disulfide–thiol exchange reaction between HA–pyridyldithiol and siRNA–SH. The remaining pyridyldithiol group was further blocked with cysteine to prevent the possible adverse effects. The obtained HA–siRNA conjugate was used for the formation of HA–siRNA/CSLN complex. The biomimetic CSLNs were prepared by reconstituting the composition of natural apolipoprotein-free LDLs.21 The HA–siRNA/CSLN complex was characterized by gel electrophoresis (GE), light dynamic scattering (DLS), and atomic force microscopy (AFM). After in vitro cytotoxicity test, target specific gene silencing tests of HA–siRNA/CSLN complex were carried out in the mouse factor VII (mFVII) expressing-HeLa cells using the mFVII siRNA. We evaluated the therapeutic gene silencing of HA–siRNA/CSLN complexes in comparison with those of HA–siRNA conjugates, siRNA complexed with commercialized siRNA transfection reagents, and siRNA/CSLN complex. The feasibility of HA–siRNA/CSLN complex was discussed for the target specific treatment of various liver diseases.
Experimental section
Materials
HA (100 kDa) was purchased from Shiseido (Tokyo, Japan). Dimethyl sulfoxide (DMSO), 1,4-diaminobutane (DAB), L-cysteine were obtained from Sigma-Aldrich (St. Louis, MO). 1-Hydroxybenzotriazole monohydrate (HOBt) was obtained from Daejung Chemicals & Metals (Shiheung, Korea). Succinimidyl-3-(2-pyridyldithio)propionate (SPDP) was purchased from Thermo Scientific (Rockford, IL). 1-Ethyl-3-[3-(dimethylamino)propyl]carbodiimide (EDC) hydrochloride was obtained from Tokyo Chemical Industry (Tokyo, Japan). Cholesteryl oleate, cholesterol hydrochloride, glyceryl trioleate (triolein), L-α-dioleoylphosphatidylethanolamine (DOPE), 3b-[N-(N′,N′-dimethylaminoethane)carbamoyl]cholesterol (DC-Chol), and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy-PEG2k] (DSPE-PEG2k) were purchased from Avanti Polar Lipids (Alabaster, AL). In vivo jetPEI and INTERFERin were purchased from Polyplus Transfection (Illkirch, France). BIOPHEN FVII kit (Ref A221304) was purchased from Aniara (West Chester, OH). The modified simFVII was obtained from ST Pharm (Seoul, Korea). The sequences were 5′-GGA ucA ucu cAA Guc uuA cdT*dT-3′ (sense) and 5′-GuA AGA cuu GAG AuG Auc cdT*dT-3′ (antisense). The 3′-end of sense strand was modified to thiol group. Small letters are for 2′-fluoro addition and * is for phosphorothioate. All reagents were used without further purification.Synthesis and characterization of HA–siRNA conjugate
HA was chemically modified with DAB and SPDP as we reported elsewhere.20 One hundred mg of HA was dissolved in 10 mL of distilled water. Two hundred mg of EDC, 135 mg of HOBt, and 200 mg of DAB were added to the HA solution. Then, the pH was adjusted to 4.8. After reaction at room temperature for 5 min, the resulting HA–DAB conjugate was dialyzed and lyophilized. The synthesized HA–DAB (10 mg) was dissolved in 1 mL of PBS (pH 7.4). SPDP at 2 molar ratios of amine groups of HA–DAB was dissolved in 1 mL of DMSO and added to the HA–DAB solution. After reaction at room temperature for 2 h, the resulting solution was dialyzed and lyophilized. The degree of modification was analysed by 1H nuclear magnetic resonance (NMR). Thiolated siRNA at 10 molar ratio of one HA chain was added to the HA–DAB–SPDP solution in PBS (pH 7.4) and reacted for 12 h. Then, L-cysteine at 2 molar ratios of 2-pyridyldithiol group in HA–DAB–SPDP was added to the HA–siRNA conjugate solution and reacted for 2 h. HA–siRNA conjugate after cysteine blocking was rapidly purified using a centrifugal filter to remove the remaining cysteine after the reaction. The final crude products were purified by the fractionation to remove free siRNA using size exclusion chromatography (SEC). The detection wavelengths were 210 nm for HA and 260 nm for siRNA. The purified HA–siRNA conjugate solution was concentrated using a speed vac (Micro-Cenvac, N-BIOTEK, Korea).Preparation and characterization of CSLN
CSLNs were prepared as we reported elsewhere.21 Briefly, cholesterol oleate (22.5 mg), triolein (1.5 mg), cholesterol (4.95 mg), DOPE (7.0 mg), DC-Chol (14.0 mg), and DSPE-PEG2k (0.05 mg) were added to 2 mL of chloroform–methanol co-solvent at a volume ratio of 2/1. Ten mL of deionized (DI) water was added. The mixture was vortexed for 2 min and subsequently sonicated for 5 min using a Sonics Vibra-Cell Sonifier VC750 equipped with a micro-tip (Newtown, CT). Then, the solvent was evaporated using a rotary evaporator (EYELA, Japan). The resulting CSLNs were dialyzed against DI water (MWCO = 10 kDa). The topological analysis of CSLNs was carried out with a tapping-mode AFM (Veeco NanoScope IV Multimode AFM, Santa Barbara, CA). The average particle size was determined by measuring the diameters of more than 20 particles on the images. The hydrodynamic size and zeta potential were analysed with a Zetasizer Nano (Malvern Instrument Co., UK), (n = 3).Preparation and characterization of HA–siRNA/CSLN complex
HA–siRNA/CSLN complexes were prepared by mixing HA–siRNA conjugate solution (1 mg mL−1) with CSLN solution (5 mg mL−1) at various w/w ratios and incubation in PBS (pH 7.4) for 15 min. The hydrodynamic volume and surface charge of HA–siRNA/CSLN complex were measured with the Zetasizer Nano (Malvern Instrument Co., UK) after dilution of the complex solution with 1 mL of PBS (pH 7.4). Then, HA–siRNA/CSLN complex at the w/w ratio of 20 was used for the following experiments.In vitro cytotoxicity and gene silencing tests
Mouse FVII (mFVII) expressing-HeLa cells were cultured in RPMI 1640 supplemented with 10% FBS, 1% antibiotics (penicillin), and 200 μg mL−1 geneticin in a 5% CO2 incubator at 37 °C. The mFVII-HeLa cells at a population of 5 × 103 were dispensed in each well of 96-well plate. After incubation for 12 h, the media were changed with fresh media containing 10% FBS and various concentrations of simFVII/CSLN and HA–simFVII/CSLN complexes. After incubation for 24 h, the media were changed with fresh media and incubated for an additional 24 h. Then, the supernatants were collected for gene silencing tests and fresh media without FBS were added. The cytotoxicity was evaluated by MTS assay. In vitro gene silencing efficiency was analyzed by measuring the concentration of mFVII enzyme in the supernatants collected from the above experiments using the BIOPHEN FVII kit. The HA–simFVII conjugates or the complex of simFVII with in vivo jetPEI or INTERFERin were prepared following the product instructions and used for the analysis of the cytotoxicity and the gene silencing efficiency. The untreated cells were used as a control (n = 3).Results and discussion
Synthesis and characterization of HA–siRNA conjugate
Fig. 1a shows a schematic illustration for the synthesis of reducible HA–siRNA conjugate. The reducible HA–siRNA conjugate was synthesized by the disulfide–thiol exchange reaction between HA–DAB–SPDP and siRNA–SH.20 The remaining pyridyldithiol groups were further blocked with cysteine for the prevention of possible adverse effects. The chemical modification degree of HA influences on the binding affinity to the HA receptors.18 Thus, the HA–DAB–SPDP conjugate was prepared to have a degree of HA modification less than 25 mol% maintaining the HA receptor binding affinity for the target specific delivery. | ||
| Fig. 1 Schematic representations for (a) the synthesis of reducible HA–siRNA conjugate and (b) the preparation of HA–siRNA/CSLN complex. | ||
Fig. 2a shows the 1H NMR spectrum of HA–DAB–SPDP conjugate. The peak at δ = 1.3–1.6 ppm corresponds to DAB, the peaks at δ = 7.0–8.5 ppm do to SPDP, and the peak at δ = 1.9–2.0 does to acetamido moiety of HA. The quantitative peak analysis confirmed the successful preparation of slightly modified HA–DAB (10 mol%)–SPDP (10 mol%). The HA–DAB–SPDP was conjugated with siRNA–SH to prepare reducible HA–siRNA conjugate with the disulfide bond which can be cleaved by glutathione in the cytosol. Then, the remaining pyridyldithiol groups of HA–siRNA conjugates were blocked with cysteine. Fig. 2b shows the 1H NMR of HA–siRNA conjugate after cysteine blocking. The peak at δ = 2.8–3.0 ppm corresponds to the grafted cysteine. The quantitative peak analysis showed that the remaining 2-pyridyldithiol group was replaced completely by cysteine.
The successful synthesis of HA–siRNA conjugate after cysteine blocking was confirmed by SEC and agarose GE. The retention time of siRNA was ca. 42 min. After conjugation, the absorbance peak of siRNA appeared at the shortened time of ca. 20 min, indicating that siRNA was clearly conjugated to HA (Fig. 3a). The bioconjugation efficiency, representing the molar ratio of siRNA conjugated with HA to the siRNA added initially, was ca. 65%. It was not almost affected by the cysteine blocking. The HA–siRNA conjugate was fractionated with SEC to have a purity more than 98% (Fig. 3b). The siRNA contents in fractionated HA–siRNA conjugate were determined by the analysis of UV-vis absorbance at 210 nm for HA and 260 nm for siRNA using the standard curves of HA and siRNA. HA–siRNA conjugates contained ca. 6 siRNA molecules per single HA chain. The UV-vis spectra with DTT treatment also indicated that there was no remaining 2-pyridyldithiol group in the HA–siRNA conjugates after cysteine blocking (Fig. 3c).
 | ||
| Fig. 3 (a) SEC, (b) agarose GE of HA–siRNA conjugate, and (c) UV-vis spectra of the HA–siRNA conjugate in the presence of DTT before or after blocking with cysteine. | ||
Preparation of cationic solid lipid nanoparticles (CSLN)
The clinically feasible delivery carrier of siRNA is highly required for its further development. Among siRNA delivery carriers, solid lipid nanoparticles (SLNs) are well-known promising lipid nanocarriers for intracellular drug delivery.22 SLNs are stabilized liposomes with a solid lipid core matrix which can solubilize lipophilic molecules with the advantages of liposomes, such as cell membrane mimicking, cell-specific interactions, and so on.23 It has been reported that SLNs were more efficiently endocytosed than liposomes due to the enhanced stability.23 Furthermore, in our previous report,23 CSLNs derived from apolipoprotein-free low LDLs, which are recognized by lipoprotein receptors in the liver like LDL receptor and remnant receptor, were used for target specific delivery of siRNA to treat liver fibrosis.23 Here, we tried to develop HA–siRNA/CSLN complex for the effective liver-specific gene silencing. As we reported previously,21 we prepared biomimetic CSLNs by reconstituting the composition of natural apolipoprotein-free LDLs using the modified emulsification and solvent evaporation method. CSLNs were composed of a hydrophobic inner core (cholesterol oleate, triolein, and cholesterol) and a surrounding surface monolayer (DC-Chol, DOPE, and DSPE-PEG2k). Because cholesteryl oleate has a high melting temperature (Tm = 52 °C), the CSLNs can be greatly stable in the body in comparison to the conventional liposomes. Cationic polar lipid DC-Chol and fusogenic lipid DOPE can provide a positive surface charge and facilitate the efficient intracellular uptake. The resulting CSLN solution is milky white in colour (Fig. 4a). AFM image showed the well-dispersed spherical morphology of CSLNs with an average diameter of 97 ± 14 nm (Fig. 4b). From the DLS analysis, the average hydrodynamic volume size was determined as 119.3 nm with a PDI of 0.218 (Fig. 4c). The ζ-potential of CSLNs was determined to be 51.1 mV (Fig. 4d). The highly positive surface charge is adequate to condense negatively charged HA–siRNA conjugate into a stable nano-sized complex.Preparation and characterization of HA–siRNA/CSLN complex
As schematically shown in Fig. 1b, HA–siRNA/CSLN complex was prepared and characterized with agarose GE, DLS, and AFM. It was compared with siRNA/CSLN complex. CSLN was mixed with siRNA or HA–siRNA conjugate at various weight ratios from 5 to 30 in PBS (pH 7.4). As shown in Fig. 5a, the migrations of siRNA and HA–siRNA conjugate were retarded above a weight ratio of 20. The siRNA/CSLN and HA–siRNA/CSLN complexes at a weight ratio of 20 were analysed by AFM and DLS. AFM analysis revealed the well dispersed siRNA/CSLN and HA–siRNA/CSLN complexes with the particle sizes of 105 ± 23.73 nm and 130 ± 24.45 nm, respectively (Fig. 5b). The average hydrodynamic sizes of siRNA/CSLN and HA–siRNA/CSLN complexes by DLS were 210 nm with a PDI of 0.165 and 303 nm with a PDI of 0.193, respectively (Fig. 6a). The relatively large particle size by DLS might be ascribed to the analysis in the hydrated state. From the results, we confirmed that siRNA and HA–siRNA conjugate could make well dispersed and nano-sized complexes with CSLNs. The surface charges were also analysed by measuring the ζ-potential value (Fig. 6b). While siRNA/CSLN complex still had a highly positive surface charge of 46 mV, HA–siRNA/CSLN complex had a slightly negative surface charge of −12 mV. The positive surface charge of siRNA/CSLN complex might trigger the nonspecific binding with blood serum components resulting in fast degradation of siRNA and bind to negatively charged cell membrane resulting in a nonspecific delivery of siRNA.4,19,20 On the other hand, the slightly negative surface charge of HA–siRNA/CSLN complex due to the presence of HA might reduce the non-specific interaction with blood serum components and enhance the target specific delivery of siRNA to the liver tissue with HA receptors. The solution stability of siRNA/CSLN and HA–siRNA/CSLN complexes was investigated at the high concentration of siRNA (0.05 mg mL−1 and 0.1 mg mL−1) in PBS (pH 7.4) to check in vivo applicability. As shown in Fig. 6c, siRNA/CSLN and HA–siRNA/CSLN complexes were highly stable at the high concentration without aggregation. We confirmed the average hydrodynamic size and surface charge of the complex were not changed significantly up to 7 days by the DLS analysis. | ||
| Fig. 6 (a) Hydrodynamic size, (b) ζ-potential analysis, and (c) photographs of siRNA/CSLN and HA–siRNA/CSLN complexes. | ||
Cytotoxicity and gene silencing efficiency
We examined the cytotoxicity (Fig. 7) and the gene silencing efficiency (Fig. 8) of siRNA/CSLN and HA–siRNA/CSLN complexes at the w/w ratio of 20 in mFVII expressing HeLa cells. We used mFVII siRNA with increasing concentration in the presence of serum. The blood coagulation factor VII (FVII) is physiologically synthesized in the liver and released into the blood.25 In our previous report,18 we confirmed the higher gene silencing efficiency of HA–siRNA/LPEI (25 kDa) complex than siRNA/LPEI complex. However, the commercial LPEI was reported to contain residual N-acyl groups, which severely reduce the transfection efficiency of nucleic acids.24 Here, we investigated the cytotoxicity and gene silencing efficiency of siRNA and HA–siRNA complexed with the commercialized transfection reagents like in vivo jetPEI and INTERFERin. In vivo jetPEI (polyplus) is a LPEI with the MW of 25 kDa optimized for the delivery of DNA and siRNA. INTERFERin is a non-liposomal cationic and amphiphilic transfection reagent developed for the delivery of siRNA. The complex with in vivo jetPEI or INTERFERin was prepared following the product instructions. The cytotoxicity was investigated by MTS assay in the mFVII expressing HeLa cells 2 days after the sample treatment. The lethal concentration 50 (LC50) values were determined to be 92 nM of siRNA/in vivo jetPEI, 127 nM of HA–siRNA/in vivo jetPEI, 50 nM of siRNA/INTERFERin, 40 nM of HA–siRNA/INTERFERin, 750 nM of siRNA/CSLN, and 585 nM of HA–siRNA/CSLN complexes. The complex with CSLN showed a remarkably higher value than those with in vivo jetPEI and INTERFERin. These results indicated the relatively high biocompatibility of CSLN as a safe delivery vehicle of siRNA.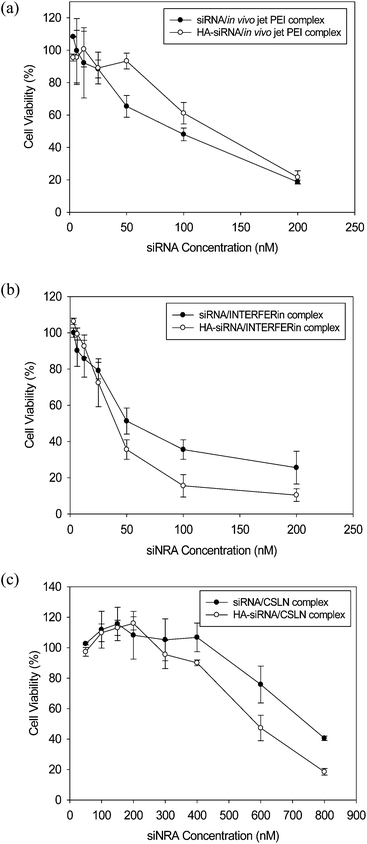 | ||
| Fig. 7 Cytotoxicity of siRNA and HA–siRNA conjugate complexed with (a) in vivo jetPEI, (b) INTERFERin, and (c) CSLN (n = 3). | ||
 | ||
| Fig. 8 Gene silencing of siRNA and HA–siRNA conjugate complexed with (a) in vivo jetPEI, (b) INTERFERin, and (c) CSLN (n = 3). | ||
The gene silencing efficiency was evaluated by measuring the mFVII concentration in the supernatants of the mFVII-expressing-HeLa cells 2 days after the sample treatment. The inhibition concentration 50 (IC50) values of complexes were determined to be 43 nM of siRNA/in vivo jetPEI, 37 nM of HA–siRNA/in vivo jetPEI, 20 nM of siRNA/INTERFERin, 8.5 nM of HA–siRNA/INTERFERin, 108 nM of siRNA/CSLN, and 60 nM of HA–siRNA/CSLN, respectively. To investigate the accurate gene silencing effect, we determined the therapeutic index (LC50/IC50) as described in Table 1. The therapeutic indexes were 2.14 of siRNA/in vivo jetPEI, 3.43 of HA–siRNA/in vivo jetPEI, 2.5 of siRNA/INTERFERin, 4.7 of HA–siRNA/INTERFERin, 6.9 of siRNA/CSLN, and 9.8 of HA–siRNA/CSLN complexes. The therapeutic indexes for the complexes with HA–siRNA conjugates were higher than those for the complexes with siRNA. The conjugation of siRNA with HA might contribute to reduce its degradation in the serum and facilitate its cellular uptake to HeLa cells with CD44 receptor for HA receptor mediated endocytosis. The delivery efficiency of siRNA or HA–siRNA conjugate was higher for the case with CSLN than INTERFERin and in vivo jetPEI. Especially, the therapeutic index of HA–siRNA/CSLN complex was statistically much higher than those of siRNA or HA–siRNA conjugate complexed with in vivo jetPEI and INTERFERin, as well as siRNA/CSLN complex. These results might be ascribed to the synergistic effect of the conjugation of siRNA with HA and biocompatible CSLN. Taken together, we could confirm the feasibility of HA–siRNA/CSLN complex for further in vivo studies and clinical applications. The HA–siRNA/CSLN complex might be effectively used to prepare a diverse siRNA complex for target specific systemic treatment of various liver diseases, such as liver fibrosis and liver cancer, by the dual targeting effect of HA and CSLN.
| Sample | LC50 | IC50 | Therapeutic index |
|---|---|---|---|
| siRNA/in vivo jetPEI | 92 ± 12 nM | 43 ± 5 nM | 2.2 ± 0.5 |
| HA–siRNA/in vivo jetPEI | 127 ± 11 nM | 37 ± 4 nM | 3.5 ± 0.4 |
| siRNA/INTERFERin | 50 ± 2 nM | 20 ± 4 nM | 2.6 ± 0.5 |
| HA–siRNA/INTERFERin | 40 ± 5 nM | 8.5 ± 1 nM | 4.7 ± 0.7 |
| siRNA/CSLN | 750 ± 28 nM | 108 ± 11 nM | 6.9 ± 0.6 |
| HA–siRNA/CSLN | 585 ± 36 nM | 60 ± 4 nM | 9.8 ± 0.2 *, ** |
Conclusion
We successfully synthesized reducible HA–siRNA conjugate for the development of HA–siRNA/CSLN complex with low cytotoxicity and high transfection efficiency for effective liver-specific gene silencing by the dual targeting effect of HA and CSLN. The formation of HA–siRNA/CSLN complex was confirmed by agarose GE, DLS, and AFM. The highly negative and flexible HA–siRNA conjugate could make a nano-sized complex with biomimetic CSLNs via electrostatic interaction. The cytotoxicity and the gene silencing efficiency of HA–siRNA/CSLN complex were assessed in mFVII expressing HeLa cells. The therapeutic index (LC50/IC50) of HA–siRNA/CSLN complex was statistically much higher than those of siRNA and HA–siRNA complexed with commercialized siRNA transfection reagents, in vivo jetPEI and INTERFERin, as well as siRNA/CSLN complex. The HA–siRNA/CSLN complex can be effectively applied as a model system for the treatment of various liver diseases, such as liver fibrosis and liver cancer, using the disease related siRNAs.Acknowledgements
This work was financially supported by the Converging Research Center Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0081871). This study was also supported by Mid-career Researcher Program through NRF grant funded by the MEST (no. 2012R1A2 A2A06045773). We greatly appreciate the financial support from SamyangBioPharm.Notes and references
- A. J. Hamilton and D. C. Baulcombe, Science, 1999, 286, 950 CrossRef CAS.
- B. L. Davidson and P. B. McCray Jr, Nat. Rev. Genet., 2011, 12, 329 CrossRef CAS PubMed.
- H. Mok, S. H. Lee, J. W. Park and T. G. Park, Nat. Mater., 2010, 9, 272 CAS.
- M. Y. Lee, S. J. Park, K. Park, K. S. Kim, H. Lee and S. K. Hahn, ACS Nano, 2011, 5, 6138 CrossRef CAS PubMed.
- E. Wagner, Acc. Chem. Res., 2012, 45, 1005 CrossRef CAS PubMed.
- A. Schroeder, C. G. Levins, C. Cortez, R. Langer and D. G. Anderson, J. Intern. Med., 2010, 267, 9 CrossRef CAS PubMed.
- A. K. Lytton-Jean, R. Langer and D. G. Anderson, Small, 2011, 7, 1932 CrossRef CAS PubMed.
- J. Soutschek, A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan and H. P. Vornlocher, Nature, 2004, 432, 173 CrossRef CAS PubMed.
- S. A. Moschos, S. W. Jones, M. M. Perry, A. E. Williams, J. S. Erjefalt, J. J. Turner, P. J. Barnes, B. S. Sproat, M. J. Gait and M. A. Lindsay, Bioconjugate Chem., 2007, 18, 1450 CrossRef CAS PubMed.
- K. Nishina, T. Unno, Y. Uno, T. Kubodera, T. Kanouchi, H. Mizusawa and T. Yokota, Mol. Ther., 2008, 16, 734 CrossRef CAS PubMed.
- C. Wolfrum, S. Shi, K. N. Jayaprakash, M. Jayaraman, G. Wang, R. K. Pandey, K. G. Rajeev, T. Nakayama, K. Charrise, E. M. Ndungo, T. Zimmermann, V. Koteliansky, M. Manoharan and M. Stoffel, Nat. Biotechnol., 2007, 25, 1149 CrossRef CAS PubMed.
- Y. L. Chiu, A. Ali, C. Y. Chu, H. Cao and T. M. Rana, Chem. Biol., 2004, 11, 1165 CrossRef CAS PubMed.
- S. A. Moschos, S. W. Jones, M. M. Perry, A. E. Williams, J. S. Erjefalt, J. J. Turner, P. J. Barnes, B. S. Sproat, M. J. Gait and M. A. Lindsay, Bioconjugate Chem., 2007, 18, 1450 CrossRef CAS PubMed.
- T. C. Chu, K. Y. Twu, A. D. Ellington and M. Levy, Nucleic Acids Res., 2006, 34, e73 CrossRef PubMed.
- S. H. Kim, J. H. Jeong, S. H. Lee, S. W. Kim and T. G. Park, J. Controlled Release, 2006, 116, 123 CrossRef CAS PubMed.
- M. Oishi, Y. Nagasaki, K. Itaka, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2005, 127, 1624 CrossRef CAS PubMed.
- J. H. Jeong, H. Mok, Y. Oh and T. G. Park, Bioconjugate Chem., 2009, 20, 5 CrossRef CAS PubMed.
- K. S. Kim, W. Hur, S. J. Park, S. W. Hong, J. E. Choi, E. J. Goh, S. K. Yoon and S. K. Hahn, ACS Nano, 2010, 4, 3005 CrossRef CAS PubMed.
- K. Park, S. W. Hong, W. Hur, M. Y. Lee, J. A. Yang, S. W. Kim, S. K. Yoon and S. K. Hahn, Biomaterials, 2011, 32, 4951 CrossRef CAS PubMed.
- K. Park, J. A. Yang, M. Y. Lee, H. Lee and S. K. Hahn, Bioconjugate Chem., 2013, 24, 1201 CrossRef CAS PubMed.
- W. H. Kong, K. Park, M. Y. Lee, H. Lee, D. K. Sung and S. K. Hahn, Biomaterials, 2013, 34, 542 CrossRef CAS PubMed.
- V. Jenning, A. F. Thünemann and S. H. Gohla, Int. J. Pharm., 2000, 199, 167 CrossRef CAS.
- W. K. Chang, Y. J. Tai, C. H. Chiang, C. S. Hu, P. D. Hong and M. K. Yeh, Int. J. Nanomed., 2011, 6, 2403 CAS.
- M. Thomas, J. J. Lu, Q. Ge, C. Zhang, J. Chen and A. M. Klibanov, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5679 CrossRef CAS PubMed.
- B. Furie and B. C. Furie, Cell, 1988, 53, 505 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |



