Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescent nitric oxide detection using cobalt substituted myoglobin†
Trevor D. Rapson*,
Helen Dacres and
Stephen C. Trowell
CSIRO Ecosystem Sciences and Food Futures Flagship, GPO Box 1700, 2601, ACT, Australia. E-mail: trevor.rapson@csiro.au; Fax: +61 2 62464000; Tel: +61 2 62464104
First published on 3rd February 2014
Abstract
We report two advances in optical biosensing of nitric oxide (NO). Firstly, we developed an improved biomolecular gated system for fluorescent transduction of heme–NO binding. Secondly, through a cobalt substitution, the detection limit for NO was decreased an order of magnitude lower than that of native myoglobin.
Nitric oxide (NO) sensors are useful for a variety of applications ranging from pollution monitoring to clinical diagnostics and biomedical research.1–3 Elevated NO levels in breath have been used as a marker for the diagnosis of airway inflammation in asthma and chronic obstructive pulmonary disease, allowing early diagnosis and intervention.4 Bioimaging of in situ NO production has allowed the elucidation of its many biological roles.2 While there are a number of NO sensors commercially available,5–8 there is a need for cheap and compact NO breath sensors to enable early detection of asthma attacks and new methods for detecting NO in vivo. Biosensors could provide a useful option for developing cheaper and more selective NO sensors.6,7
Heme proteins are ideal recognition elements for gases such as oxygen and NO. Soluble guanylate cyclase (sGC), a heme protein, can bind nitric oxide in preference over other gases such as oxygen.9 While a number of heme proteins have been tested for their suitability as NO optical sensors, they do not have the sensitivity provide by techniques such as chemiluminescence or electrochemistry. Furthermore, immobilisation of heme proteins can adversely affect both their response and reversal times.10–14
In order for heme proteins to be effectively used as optical biosensors, their sensitivity needs to be increased and limits of detection (LOD) needs to be reduced. In this study, we sought to improve the sensitivity of heme protein based optical transducers using two approaches. The first involves developing an efficient fluorescent method to detect NO binding to heme proteins. The second involves using a metal substituted myoglobin, cobalt myoglobin (CoMb) to decrease the LOD for NO.
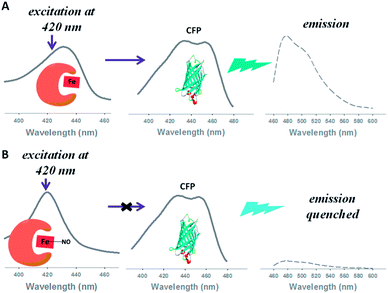
We employed a relatively new fluorescent method known as biomolecular gating.15 In this approach, the fluorescence intensity of a fluorophore is modulated by the differing amount of light absorbed by a heme protein in the NO-bound or NO-free state. The heme protein effectively acts as a wavelength selective filter, limiting the exciting light reaching the fluorophore (Scheme 1).15
In the first reported biomolecular gating system, Strianese et al.15 employed two heme proteins; cytochrome c peroxidase and myoglobin to measure NO and O2 binding respectively. Fluorescein immobilised on a nitrocellulose membrane, excited at 450 nm, was used as the fluorophore. Although the authors were able to convert absorption changes into either an increase or decrease in fluorescence, quantitative analysis of gas binding was not reported.
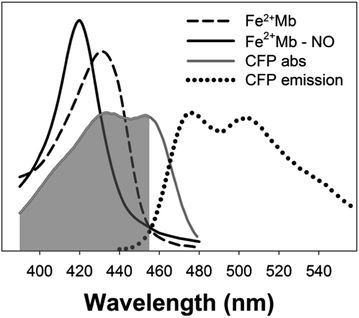
The key to an efficient molecular gating system is to optimise the overlap between the excitation band of the fluorophore and one or more of the absorption bands of the gas binding protein.15 In order to improve the efficiency of biomolecular gating we chose a fluorophore with a large spectral overlap with heme proteins in the 400–430 nm region. Cyan fluorescent protein (CFP) with absorption maxima at 435 nm and 455 nm has large spectral overlap with reduced, oxidised and nitric oxide bound myoglobin (Fig. 1). Fluorescent proteins in biosensors are also advantageous to small molecular dyes such as fluorescein as they have greater photostability.16 A further modification to the procedure reported by Strianese et al.15 was to have both the fluorophore and NO binding protein in solution rather than immobilising the fluorophore on a membrane.
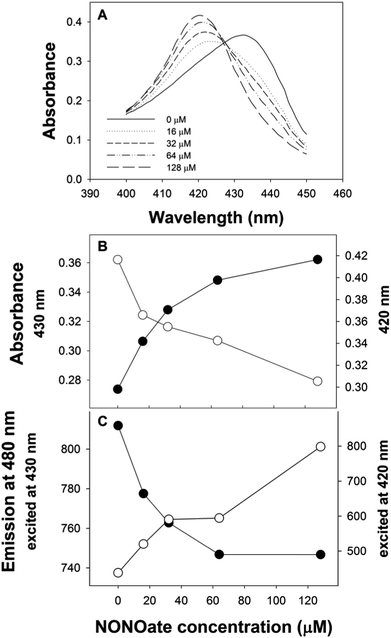
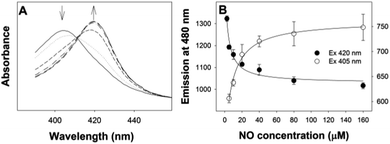
With CFP as the fluorophore, we used excitation wavelengths of 400–430 nm. The changes in the position of the Soret peak with increasing dissolved NO concentration (generated by diethylamine NONOate, see ESI†) were converted into quantitative fluorescence changes (Fig. 2). When exciting at 420 nm, the increase in absorption at 420 nm of myoglobin with NO (Fig. 2B) led to a decrease in CFP emission at 480 nm (Fig. 2C). Conversely, the decrease in myoglobin absorption at 430 nm (Fig. 2B) led to an increase in CFP fluorescence when exciting at 430 nm (Fig. 2C). No fluorescence changes were seen with the addition of NO to CFP.
The biomolecular gating system with myoglobin and CFP was optimised for the “gating” effect of myoglobin. Optimal concentrations of 15 μM and 0.65 μM for myoglobin and CFP respectively were determined. Using the optimised conditions, there was ∼85% decrease in the initial fluorescence signal with excitation at 420 nm, substantially greater than the 20% decrease previously reported with fluorescein.15
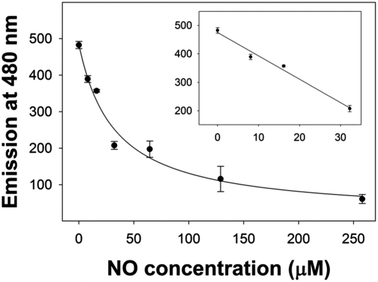
Initially, both 420 and 430 nm were used as excitation wavelengths. However, excitation at 420 nm was found to give improved reproducibility and larger changes in emission compared to excitation at 430 nm. Using an excitation wavelength of 420 nm, with myoglobin, a linear region between 0–30 μM NO was obtained and it was possible to determine the dissociation constant (Kd) of NO binding (Fig. 3).
We hypothesised that an iron to cobalt substitution could improve the limit of detection for NO. CoMb (where iron protoporphyrin IX has been substituted with cobalt protoporphyrin IX) has been used previously to study the function of oxygen binding heme proteins such as hemeoglobin and myoglobin. CoMb has a number of properties which could make it ideal for use as a NO sensor. Firstly, Co2+Mb has a lower oxygen affinity (50–100 times larger partial pressure of oxygen, p0.5) than native myoglobin,17,18 while showing an intrinsically greater reactivity toward NO.19 Furthermore, CoMb does not bind carbon monoxide,18 and has decreased affinity for nitrates and nitrites.20
Laverman and Ford carried out mechanistic studies of NO reactions with iron and cobalt porphyrin complexes. They reported a larger formation constant for Co2+ porphyrins (1.3 × 1013 M−1) compared to Fe2+ porphyrins (2.4 × 1012 M−1).21 No one, however, has investigated the suitability of CoMb for use as an optical NO biosensor. We, therefore used our biomolecular gating system, to compare NO binding properties of FeMb and CoMb.
The Soret peak of Co2+Mb shifts from 405 nm to 420 nm on NO binding (Fig. 4A). Using CoMb, quantitative analysis of NO binding was carried out at both 405 nm and 420 nm. We observed that Co2+Mb was less readily oxidised than Fe2+Mb giving data with lower variance and improved reproducibility for Co2+Mb when tested under both aerobic and strict anaerobic conditions. Again, 420 nm is the preferred excitation wavelength as we observed less scatter compared to excitation at 405 nm (Fig. 3B).
In order to quantitatively compare the suitability of FeMb and CoMb for use in NO sensing, we determined their Kd and LOD when excited at 420 nm. The same NONOate solutions were used to allow direct comparison. FeMb had a Kd of 32 ± 10 μM while the Kd of CoMb was three times lower at 10 ± 2 μM. There was a significant reduction in the LOD of dissolved NO from 10 μM for FeMb to 1 μM for CoMb.
This ten-fold decrease in the limit of detection for dissolved NO obtained using CoMb demonstrates that Co substitution could improve the sensitivity of heme protein-based NO sensors. The use of metal substitution has been reported for other heme proteins. Recently, Marletta and co-workers developed an innovative system where metal substitution is carried out during protein expression rather than traditionally used denaturing conditions.22 Therefore, the use of metal substituted heme proteins in biosensors is becoming more convenient and reliable.23
In addition to the decrease in the limit of detection, a cobalt substitution introduces a number of other useful properties. The lower affinity of CoMb for carbon monoxide and nitrates,18,20 is potentially an advantage if the sensor were to be used for environmental testing.
The limit of detection obtained with the CoMb/CFP molecular gated system is comparable to the best heme fluorescent system reported to date of 1 μM for soluble guanylate cyclase (sGC) (Table 1),10 albeit the FRET sGC system was immobilised. A limit of detection of 1 μM NO in solution is equivalent to a concentration of 30 ppb, this is within the useful range for measurement of NO in breath. In this study we have been measuring dissolved NO generated by diethylamine NONOate to compare FeMb and CoMb, future work will involve measuring NO from gas mixes and ultimately breath samples in order to develop a NO biosensor for the early detection of asthma attacks.
| Heme receptor | Transduction method | Limit of detection | Reference |
|---|---|---|---|
| a Thermoanaerobacter tengcongensis heme nitric oxide binding protein.b Förster resonance energy transfer. n.d. – not determined. | |||
| Cytochrome c | Absorbance | 1 ppm | 24 |
| Cytochrome c′ | Absorbance | 10 μM | 12 |
| Myoglobin | Absorbance | n.d. | 25 |
| Tt H-NOXa | Absorbance | 0.3 μM | 14 |
| Cytochrome c′ | FRETb | 8 μM | 11 |
| Soluble guanylate cyclase | FRETb | 1 μM | 10 |
| Cytochrome c peroxidase | FRETb | n.d. (Kd = 10 μM) | 13 |
| Cytochrome c peroxidase | Biomolecular gating | n.d. | 15 |
| Myoglobin | Biomolecular gating | 1 μM (30 ppb) | This work |
It may be possible to extend the use of biomolecular gated detection to in vivo tissue imaging. Fluorescent proteins have been widely used for biological imaging,16 therefore using a biomolecular gating system with heme and fluorescent proteins, we will determine if it is possible to detect NO production in vivo.
We used cobalt substituted myoglobin for this study as cobalt substitutions in the protein have been well characterised. There are a number candidate heme proteins which could be used in a NO sensor, such as Tt H-NOX (Table 1).9,14,23 The findings presented here suggest that it could be worthwhile investigating cobalt substitutions in a range of heme proteins to determine if lower detection limits can be obtained.
Conclusions
Using CFP and myoglobin we have developed an improved molecular gating system which allowed quantitative analysis of NO binding. The LOD of a myoglobin nitric NO detection system was improved from 10 μM to 1 μM through an iron to cobalt substitution. These are promising advances in the development of optical biosensors for nitric oxide.Notes and references
- S. M. Cristescu, D. Marchenko, J. Mandon, K. Hebelstrup, G. W. Griffith, L. A. J. Mur and F. J. M. Harren, Appl. Phys. B, 2012, 110, 203 CrossRef.
- T. Nagano and T. Yoshimura, Chem. Rev., 2002, 102, 1235 CrossRef CAS PubMed.
- N. Kumar, V. Bhalla and M. Kumar, Coord. Chem. Rev., 2013, 257, 2335 CrossRef CAS PubMed.
- A. F. Gelb, P. J. Barnes, S. C. George, F. L. M. Ricciardolo, G. DiMaria and N. Zamel, J. Breath Res., 2012, 6, 047101 CrossRef PubMed.
- R. Dweik, P. Boggs, S. Erzurum and C. Irvin, Am. J. Respir. Crit. Care Med., 2011, 184, 602 CrossRef CAS PubMed.
- C. I. L. Justino, T. A. Rocha-Santos, A. C. Duarte and T. A. Rocha-Santos, Trends Anal. Chem., 2010, 29, 1172 CrossRef CAS PubMed.
- S. Griveau and F. Bedioui, Anal. Bioanal. Chem., 2013, 405, 3475 CrossRef CAS PubMed.
- S. Jiang, R. Cheng, X. Wang, T. Xue, Y. Liu, A. Nel, Y. Huang and X. Duan, Nat. Commun., 2013, 4, 1 Search PubMed.
- E. R. Derbyshire and M. A. Marletta, Annu. Rev. Biochem., 2012, 81, 533 CrossRef CAS PubMed.
- S. L. Barker, Y. Zhao, M. A. Marletta and R. Kopelman, Anal. Chem., 1999, 71, 2071 CrossRef CAS.
- S. L. Barker, R. Kopelman, T. E. Meyer and M. A. Cusanovich, Anal. Chem., 1998, 70, 971 CrossRef CAS.
- D. J. Blyth, J. W. Aylott, J. W. Moir, D. J. Richardson and D. A. Russell, Analyst, 1999, 124, 129 RSC.
- M. Strianese, F. De Martino, V. Pavone, A. Lombardi, G. W. Canters and C. Pellecchia, J. Inorg. Biochem., 2010, 104, 619 CrossRef CAS PubMed.
- E. M. Boon and M. A. Marletta, J. Am. Chem. Soc., 2006, 128, 10022 CrossRef CAS PubMed.
- M. Strianese, A. Varriale, M. Staiano, C. Pellecchia and S. D'Auria, Nanoscale, 2011, 3, 298 RSC.
- N. C. Shaner, P. A. Steinbach and R. Y. Tsien, Nat. Methods, 2005, 2, 905 CrossRef CAS PubMed.
- T. Yonetani, H. Yamamoto and G. Woodrow, J. Biol. Chem., 1974, 682 CAS.
- B. M. Hoffman, Proc. Natl. Acad. Sci. U. S. A., 1970, 67, 637 CrossRef CAS.
- Y. Kholodenko, E. A. Gooding, Y. Dou, M. Ikeda-Saito and R. M. Hochstrasser, Biochemistry, 1999, 38, 5918 CrossRef CAS PubMed.
- J. L. Heinecke, J. Yi, J. C. M. Pereira, G. B. Richter-Addo and P. C. Ford, J. Inorg. Biochem., 2012, 107, 47 CrossRef CAS PubMed.
- L. E. Laverman and P. C. Ford, J. Am. Chem. Soc., 2001, 123, 11614 CrossRef CAS PubMed.
- J. J. Woodward, N. I. Martin and M. A. Marletta, Nat. Methods, 2007, 4, 43 CrossRef CAS PubMed.
- M. B. Winter, E. J. McLaurin, S. Y. Reece, C. Olea, D. G. Nocera and M. A. Marletta, J. Am. Chem. Soc., 2010, 132, 5582 CrossRef CAS PubMed.
- J. W. Aylott, D. J. Richardson and D. A. Russell, Chem. Mater., 1997, 9, 2261 CrossRef CAS.
- D. J. Blyth, J. W. Aylott, D. J. Richardsonb and D. A. Russella, Analyst, 1995, 120, 2725 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ra47658h |
| This journal is © The Royal Society of Chemistry 2014 |