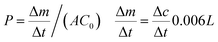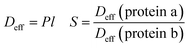Open Access Article
Open Access ArticleProtein separation performance of self-assembled block copolymer membranes†
J. Hahna,
J. I. Clodt*a,
V. Filiza and
V. Abetzab
aHelmholtz-Zentrum Geesthacht, Institute of Polymer Research Max-Planck-Str.1, 21502 Geesthacht, Germany. E-mail: juliana.clodt@hzg.de; Fax: +49 4152 872499; Tel: +49 4152 872472
bUniversity of Hamburg, Institute of Physical Chemistry, Grindelallee 117, 20146 Hamburg, Germany
First published on 17th January 2014
Abstract
A comprehensive study of the separation performance of highly porous self-assembled integral asymmetric block copolymer membranes was carried out. Four different polystyrene-b-poly(4-vinylpyridine) (PS-b-P4VP) diblock copolymers were used to prepare membranes with pore sizes increasing with the molecular weight of the polymers. The pore sizes vary from 17 nm to 53 nm. Clean water fluxes and the adsorption of lysozyme, myoglobin, haemoglobin, catalase and ferritin on the membranes were studied. Diffusion rates of the proteins were examined and selectivities are discussed. The membranes show promising results regarding the separation of different proteins in the range of around four to twelve nm. The characteristics of PS-b-P4VP diblock copolymer membranes were compared with a commercially available polycarbonate track-etched membrane.
Introduction
Ultrafiltration membranes (UF) provide huge potential for (bio)molecule separation,1,2 controlled-long term release,3 and water purification.4–6 The technology combines low energy consumption, environmental friendly conditions and can be easily combined with further technologies or be used in continuous operations.2,7–9 However, the performance of common commercially available UF is limited in separation and flux of biomacromolecules like proteins due to huge deviations in pore size, low surface porosity, the thickness of the effective separation layer,9 or a tendency to adsorb proteins.10–13 Therefore a lot of effort is made to develop optimized membranes with uniform pore sizes in the nanometer scale.14About 40 years ago, polycarbonate track-etched (PCTE) membranes with randomly distributed pores and sharp size distributions became commercially available. Nowadays, these membranes are common with pore sizes between 10 nm to 10 μm. PCTE membranes provide a good scalability while their very low porosity and limited pore uniformity, due to merging of pores, restrict their performance in separation applications. Further inorganic membranes made of aluminum anodic oxide (AAO) or mesoporous silica (MPS) provide nearly uniform pore sizes. These membranes are characterized by pore sizes of 10 nm to 200 nm for AAO and 2 nm to 10 nm for MPS and high porosities. Nevertheless these membranes are hardly scalable and very brittle.9,15
Due to the huge potential of nanoporous materials with uniform pore sizes, several studies were performed to develop improved membrane structures, to figure out and to understand their separation performance, especially for proteins.15–17 In this context the passive diffusion of proteins through membranes is a versatile method for protein separation or long-term release, since the avoidance of physical or chemical stresses protects against denaturing of the proteins.3
The combination of self-assembly of amphiphilic block copolymers (S) and the non-solvent induced phase separation process (NIPS) allows preparing integral-asymmetric membranes with highly ordered, hexagonally arranged pores in a fast one-step process.18 These membranes show a thin selective layer on top consisting of hexagonally arranged cylinders merging in a sponge-like substructure underneath. Polystyrene-block-poly(4-vinylpyridin) (PS-b-P4VP) was the first block copolymer used to prepare the desired membrane structures and is therefore well-examined regarding its membrane formation. In the last years a lot of progress was achieved in membranes fabrication using SNIPS (a combination of self-assembly and the non-solvent induced phase separation process) including e.g. the expansion on various di- and triblock copolymer systems,19–23 the investigation of the influence of additives,24–26 and of the structure formation process.27–29 This proves a high adaptability of the sensitive and fascinating process and its resulting membrane structures leading to a further increase of their range of application. Our recent results show that the pore sizes of PS-b-P4VP membranes are tunable in the range of 20 nm to 70 nm.30 The pores of the membranes mainly downsize by decreasing the molecular weights of PS-b-P4VP used for the corresponding membrane casting solutions. The potential of isoporous PS-b-P4VP membranes for the separation of biomolecules was pointed out previously in diffusion experiments using membranes with pore sizes of 34 nm.31 Bovine serum albumin (BSA) and globulin-γ(IgG) could be effectively separated. Furthermore the membrane allowed the selective separation of similarly sized proteins based on charge effects, as stated for the separation of BSA and bovine hemoglobin. After quaternization of the membrane it was possible to separate these proteins effectively by varying the pH due to their different isoelectric points.
Self-assembled PS-b-P4VP membranes under investigation with pore sizes of several tens of nanometers combine a sharp pore size distribution, high porosity and their production is easily up-scalable. This is an ideal basis for an excellent separation performance, in particular with respect to commercially available isoporous polymeric membranes. To the best of our knowledge up to now no comprehensive investigation of the performance of PS-b-P4VP membranes and the influence of various pore sizes was carried out. In this work we are looking in detail on the clean water flux over 40 h, the adsorption of proteins and the separation selectivity of PS-b-P4VP membranes with various pore sizes between five model proteins in diffusion experiments.
Experimental
PS-b-P4VP diblock copolymers
Tetrahydrofuran (THF) was ordered from Th. Geyer. Styrene and 4-vinylpyridine (4VP) were ordered from Sigma-Aldrich and purified as per description. THF was purified by successive distillation from potassium under purified argon atmosphere. Styrene was stirred with dibutylmagnesium (MgBu2). 4VP was distilled once under reduced pressure from calcium hydride (CaH2) and twice from ethylaluminium dichloride. PS-b-P4VP was synthesized via sequential anionic polymerization at −67 °C in THF. The polymerization of styrene was initiated with sec-butyl lithium (sec-BuLi). After 4 h, 4VP was added and the solution was stirred for another 16 h. The polymerization was quenched with degassed methanol. After partial removal of THF under reduced pressure, the polymer was precipitated in water. The composition of the block copolymer was determined by 1H-NMR spectroscopy. All 1H-NMR measurements were performed on a Bruker Advance 300 NMR spectrometer at 300 MHz with internal standard [tetramethylsilane (TMS)] using chloroform (CDCl3) as a solvent. Molecular weights of the PS precursors and polydispersities were determined by gel permeation chromatography (GPC). The number averaged molecular weights of the block copolymers were calculated from the number averaged molecular weight of the PS precursor obtained by GPC and the composition obtained by 1H-NMR. GPC measurements were performed at 50 °C in dimethylacetamide (DMAC) using 3 μ PSS SDV gel columns at a flow rate of 1.0 mL min−1 (VWR-Hitachi 2130 pump). A Waters 2410 refractive-index detector (λ = 930 nm) and a PS calibration curve were used.Protein solutions
Lysozyme (Lys) from chicken egg white, myoglobin (Myo) from equine skeletal muscle, haemoglobin (Hem) from bovine blood, catalase (Cat) from bovine liver and ferritin (Fer) from equine spleen were purchased from Sigma Aldrich. Sodium chloride, potassium dihydrogenphosphate and disodium hydrogenphosphate dodecahydrate used for phosphate buffered saline (PBS) solutions were purchased from Sigma Aldrich. Protein concentrations were determined using the UV-Vis spectrophotometer Genesys 10S (Thermo Scientific) at a wave length of 280 nm and 409 nm, respectively.Membranes used in this work
The isoporous diblock copolymer membranes used in this work were prepared according to a procedure recently published by Rangou et al.30 Therefore PS-b-P4VP diblock copolymers synthesized as described above were dissolved in a solvent mixture of THF and DMF. The solutions were cast on a polyester nonwoven support using a casting machine. The films were left for a certain time on air before immersing them in water. The membranes were dried at 60 °C under reduced pressure before use.The commercial polycarbonate track-etched membranes (PC30) used in this work were purchased from Pieper Filter GmbH, supplier number PCN3CP04700, pore sizes 30 nm, porosity ∼0.4 as stated by supplier.
Scanning electron microscopy (SEM) of the membranes was carried out on a LEO Gemini 1550 VP at a voltage of 3 kV or 5 kV. The samples were coated with 2.0 nm platinum. The cross-sections of the membranes were dipped in iso-propanol, frozen in liquid nitrogen and cracked. Average pore size values were determined using the software analySIS (Olympus) on basis of the SEM results. ImageJ 1.46 (Wayne Rasband, National Institute of Health, USA) was used to determine the porosity of our membranes.
Water-flux experiments
Water flux measurements were performed in dead-end mode using a home-made automatic testing device at trans-membrane pressures of 2.0 bar to 2.3 bar at room temperature. The volume was measured gravimetrically every 15 and 30 min for 40 h, respectively. The actual pressure was recorded as well. The effective membrane area was 1.77 cm2. These studies were conducted employing demineralized water with an electrical conductivity of ≈0.055 μS cm−1.Adsorption experiments
Adsorption experiments adjusted to a known procedure32 were carried out with five different proteins of different hydrodynamic diameters and isoelectric points: values for hydrodynamic diameters (Dh) and isoelectric points (IEP) as stated by supplier; Lys (Dh = 3.8 nm, IEP = 11.4), Myo (Dh = 4.4 nm, IEP = 7.2), Hem (Dh = 6.4 nm, IEP = 7.1), Cat (Dh = 10.2 nm, IEP = 5.4) and Fer (Dh = 12.0 nm, IEP = 7.2). 2 mL of a 1 g L−1 solution of the proteins in PBS buffer solution (10 mM PBS, 9 wt% of NaCl in demineralized water) at different pH were placed on a membrane disc of 2 cm in diameter in a small closed vial. The vials were shaken at 90 rpm for 24 h at 25 °C to reach equilibrium. The membranes were rinsed two times with 2 mL of the PBS buffer solution for 10 minutes. The protein adsorption on the membrane was calculated as follows:where m0 is the mass of the protein in the solution before the adsorption experiment, m1 is the mass of the protein in solution after the adsorption experiment and mw1 and mw2 are the masses of the protein in the washing solutions and A is the membrane area. The concentration of the protein solution where measured by UV as stated before.
Diffusion experiments
A side-by-side diffusion cell (PermeGear) was used to study the diffusion of five model proteins through our membranes. The system consists of two chambers with 6 mL volumes, namely the donor (right chamber) and the receptor compartment (left chamber). The orifice diameter allows an accessible membrane area of 2.04 × 10−4 m2 (effective diameter: 16.1 mm). Each chamber is equipped with a magnetic stirrer to allow continuous mixing and avoid concentration gradients in the chamber itself. The effect of stirring speed on the diffusion through the membrane was studied at 100 rpm, 200 rpm, and 300 rpm. Otherwise the stirring speed was kept constant to 100 rpm. The system is constantly heated to 37 °C. PBS with low ionic strength of 10 mM and a pH of 5.4, 7.4 and 11.8, respectively, was used as diffusion medium. The buffer was prepared using Na2HPO4 × 12H2O, KH2PO4, NaCl, and Millipore water. The pH was adjusted by adapting the proportions between the salts or by adding NaOH, respectively. The buffer was used within five days after preparation.Initially the membrane is placed between the two chambers exposing the ordered surface structure towards the donor chamber. The system is tightly sealed using a cell clamp. Both chambers are filled with 6 mL of the diffusion medium at least 16 hours before the diffusion experiment is started. This enables a good wetting and swelling of the membranes. To start the experiments both cells are emptied. The receptor cell is filled again with 6 mL PBS, while the donor cell is filled with 6 mL protein solution (0.5, 1.0, and 2.0 mg protein per mL PBS of the respective pH). Samples of the receptor medium were taken after 15 min, 1 h, 2 h, 4 h, 6 h, 8 h, 24 h, 26 h, 28 h, and 30 h. Therefore the solution in the receptor cell was completely removed and refilled with fresh PBS. After 24 h the receptor compartment was rinsed once with PBS buffer to remove residual proteins. After 30 h the solution in the donor compartment was removed for further analysis. Each diffusion measurement was performed at least three times.
Protein concentrations were determined by UV within one hour after sample collection. The flux J of the proteins through the membrane can be calculated based on the law of Fickian diffusion. Therefore the permeability coefficient can be obtained from the slope of the diffusant concentration C versus time t (1 h to 24 h).33 The error bars drawn in the diagram correspond to the maximum deviation from the mean value
The effective diffusion coefficient Deff of proteins was calculated according to Lu et al.34–36 and allows determining the theoretically separation selectivity S based on our single protein diffusion experiments
Results and discussion
Membranes used in this work
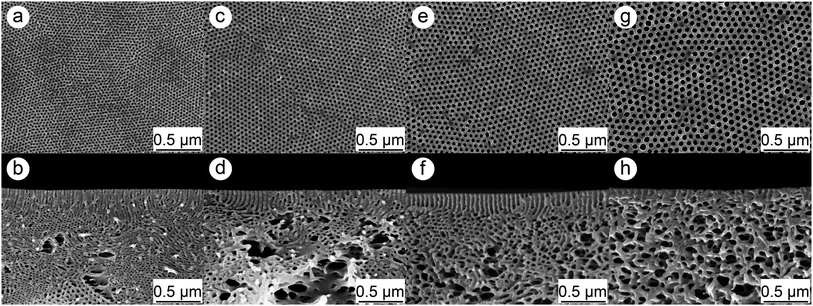
In order to obtain membranes with different pore sizes four different PS-b-P4VP diblock copolymers were used in this work. Therefore the previous published procedure37 was adjusted to develop membranes with pore sizes smaller than 20 nm. The molecular characteristics of the polymers and resulting geometrical feature of the membranes are listed in Table 1 in which the numbering of the membranes is according to their pore sizes. Fig. 1 depicts SEM image of the surfaces and cross-sections of the membranes.| PS-b-P4VP | Mn [kg mol−1] | PS [wt%] | P4VP [wt%] | Pore diametera [nm] | Average porosity [%] | Average membrane thickness [μm] | Membrane numbering |
|---|---|---|---|---|---|---|---|
| a The numeric value is attributed to the average pore size followed by the ± standard deviation. | |||||||
| PS83P4VP1776 | 76 | 83 | 17 | 17 ± 2 | 13.6 | 68 | M17 |
| PS83.7P4VP16.3113 | 113 | 83.7 | 16.3 | 23 ± 2 | 18.7 | 56 | M23 |
| PS81P4VP19160 | 160 | 81 | 19 | 34 ± 4 | 25.8 | 48 | M34 |
| PS80P4VP20252 | 252 | 80 | 20 | 53 ± 5 | 31.5 | 44 | M53 |
| Commercial polycarbonate track-etched membrane | 30 | 0.4 | 7 | PC30 | |||
Clean water-flux experiments
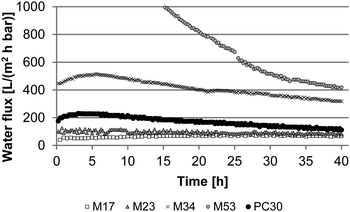
Clean water fluxes were measured over 40 hours for four different PS-b-P4VP diblock copolymer membranes (M17, M23, M34, and M53) and one commercially available polycarbonate track-etched membrane (PC30). The results are shown in Fig. 2.As expected the fluxes increase with the pore size of the PS-b-P4VP membranes. The flux declines over 40 h were calculated and are listed in Table 2. For M17, M23 and M34 the water flux values are quite stable throughout the measurement leading to small flux declines below 30%. The negative flux decline value for M17 could be caused by some small polymer particles inside the raw membrane which are pressed out during the measurement leading to an increased flux after some time. Another explanation is that the deviance of the water flux values is bigger for M17 since the values are quite small leading to a higher relative error during the measurement. The flux decline increases with the pore size of the PS-b-P4VP block copolymer membranes.
| Membrane | Initial flux [L m−2 h−1 bar−1] | Flux after 40 h [L m−2 h−1 bar−1] | Flux decline [%] |
|---|---|---|---|
| M17 | 55 | 67 | −22 |
| M23 | 101 | 90 | 11 |
| M34 | 432 | 321 | 25 |
| M53 | 1465 | 418 | 72 |
| PC30 | 198 | 111 | 44 |
Interestingly the flux decline is much higher for M53 than for the other membranes. The initial flux of this membrane was 1465 L m−2 h−1 bar−1. After 40 h it seems to approximate a stable state with a flux value of around 400 L m−2 h−1 bar−1. The huge flux decline of M53 could be caused by an increased swelling of the P4VP chains that are in this case quite longer (∼50 kg mol−1) compared to the other membranes (M34 ∼30 kg mol−1, M23 ∼18 kg mol−1, and M17 ∼13 kg mol−1). Furthermore it has to be mentioned that the water fluxes over 40 h for M34 and M53 could not be measured continuously since our measurement set up can only measure up to a limit of 4 L water. The water fluxes for M34 and M53 are so high that the overall volume of water is more than 4 L leading to interval measurements. During the measurement breaks the membranes are kept wet in the system over night. Therefore the higher flux declines for M34 and M53 could also be generated by an increased fouling on the surface due to the longer times the membranes are kept in water. An increased amount of bacteria was observed for M34 to M53 after flux measurements as shown in Fig. S1.†
Adsorption experiments
A detailed analysis of the adsorption of different proteins on isoporous diblock copolymer membranes was necessary for this study. On the one hand adsorption of proteins on membranes is correlated to their fouling behavior which is important when it comes to application. On the other hand the adsorption of proteins on the membrane surface and substructure reduces the effective pore diameters and further lead to partial or full pore blocking.35 The adsorption was tested both at physiological pH 7.4 and at the IEP of the proteins. The latter one is important because the adsorption can be different when the overall charge of the proteins are neutral (at IEP), positive (pH < IEP) or negative (pH > IEP) due to electrostatic interaction with the membrane surface. For Myo, Hem and Fer the IEP is close to pH 7.4. Therefore no additional measurements were carried out. The results of the adsorption measurements are depicted in Fig. 3 and 4.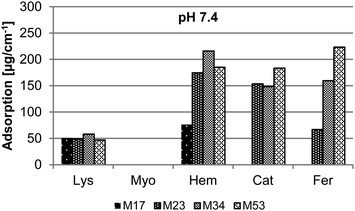 | ||
| Fig. 3 Adsorption of Lys, Myo, Hem, Cat, and Fer in PBS buffered solution (pH 7.4) on isoporous diblock copolymer membranes M17, M23, M34, and M53. | ||
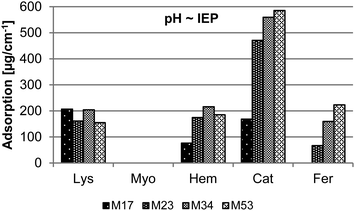 | ||
| Fig. 4 Adsorption of Lys, Myo, Hem, Cat, and Fer in PBS buffered solution (pH ∼ isoelectric point of the protein) on isoporous diblock copolymer membranes M17, M23, M34, and M53. | ||
At pH 7.4 the amount of adsorption of Lys, Myo, Hem, and Cat concerning the different membranes M23, M34, and M53 is independent of the membrane pore size and polymer used for the membrane formation, respectively. Small deviation in the absorption value may be caused by the error of the method and UV system included. Interestingly Myo is the only protein without any measurable adsorption on the membrane surface. In contrast to the other proteins the adsorption of Fer increases with the molecular weight of the polymer, respectively the pore size of the membrane. For the biggest proteins, Fer and Cat, no adsorption was observed at pH 7.4 on M17. In this case the effective pore diameters are too small for the proteins to allow any or huge amounts of proteins to reach the substructure of the membrane and adsorb there. Also for Hem the adsorption is considerably lower on the membrane with the smallest pore size M17 corresponding to the results observed for Cat and Fer.
At pH ∼ IEP the adsorption has the same tendency as for pH 7.4 when the different membranes are compared. For Lys and Cat (compare also Fig. S2†) the adsorption is much higher at neutral state (IEP) than at pH 7.4, although Lys is positively and Cat negatively charged at pH 7.4. The repulsive interaction between the proteins themselves seems to be too high to let the proteins attach to the membrane surface. In case of Cat an adsorption even for the membrane with the smallest pores M17 was observed, but the value of adsorption is still much lower than for the other membranes.
For the commercial track-etched membrane PC30 the adsorption was tested as well. Here it was not possible to detect any adsorption with our method. The supplier specifies a very low adsorption for these types of membranes which is in accordance with our measurements. Compared to PS-b-P4VP membranes, track-etched membranes do not have a spongy substructure and differ considerably in the chemical structure. On the one hand, their decreased adsorption could be due to the smaller overall surface of the polymer just as the smaller thickness of PC30 compared to our membranes. On the other hand, we assume that their functional surface groups allow less attractive interactions between the membrane and the proteins.
Diffusion experiments
In order to investigate the separation performance of our membranes, diffusion experiments with proteins were carried out. Therefore we will provide a closer look on the effect of the pore size.Besides the characteristics of the membrane, the protein and their interaction, the diffusion of proteins is affected by various measuring parameters. Some of them will be discussed as follows.
First, the ionic strength significantly affects the electrostatic double layer thickness (Debye length) of proteins and membrane surface areas. Previous studies carried out that an ionic strength of 10 mM allows combining high separation selectivity and high flux using PS-b-P4VP membranes and proteins with a hydrodynamic diameter of about 6.8 nm. These conditions lead to a Debye length of about 3.3 nm still having a significant impact especially when small protein and pore sizes are considered.31
In the following the influence of the stirring speed and the initial concentration in the donor chamber will be examined. In these studies Myo was used as a model protein, since it does not show any adsorption to PS-b-P4VP membranes. Size selective separation by diffusion of different proteins will be discussed for M17, M23, M34, M53 including the influence of pH and pore sizes. Furthermore the results are compared with the performance of the commercial membrane PC30.
Influence of stirring speed
The mass transport by diffusion through membranes is hindered by a developing laminar boundary layer influencing the diffusion coefficient Deff. The additional resistance on the surface of the membranes and further concentration gradients in the receptor chamber can be minimized due to a sufficient mixing. The influence of the stirring speed on the mass transport was determined. Therefore the stirring speed in diffusion experiments using M53 and 1 mg mL−1 Myo was varied between 100 rpm, 200 rpm, and 300 rpm. Further parameters were kept constant. The results depicted in Table 3 show, that the diffusion coefficient is slightly increasing with rising stirring speed. This effect can be attributed to the reduction of the laminar boundary layer on the surface of the membrane. For each speed step itself the difference is within the range of error. As expected, the diffusion profiles provided in the ESI Fig. S3† show that the diffusion rate increases. For our purpose we assumed a stirring speed of 100 rpm to be sufficient since above 100 rpm the stirrers in our measuring device became more and more unstable increasing the risk of membrane damage.| Stirring speed [rpm] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | Conc. Myo [mg mL−1] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] |
|---|---|---|---|---|---|
| 100 | 67.4 | 296.7 | 0.5 | 73.4 | 322.8 |
| 200 | 74.7 | 328.7 | 1.0 | 67.4 | 296.7 |
| 300 | 82.0 | 360.7 | 2.0 | 71.6 | 315.0 |
| PC30 | M17 | M23 | M34 | M53 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | P [10−6 cm s−1] | Deff [10−9 cm2 s−1] | |
| Lys | 32.6 | 221.7 | 44.3 | 248.3 | 66.2 | 317.6 | 68.8 | 302.7 | ||
| Myo | 141.1 | 98.8 | 41.3 | 280.5 | 44.6 | 249.3 | 58.8 | 282.3 | 67.4 | 296.7 |
| Hem | 8.8 | 59.6 | 10.3 | 57.9 | 20.7 | 99.1 | 23.0 | 101.1 | ||
| Cat | 75.1 | 52.6 | 11.6 | 78.8 | 15.1 | 84.8 | 25.0 | 120.1 | 34.4 | 151.5 |
| Fer | 20.7 | 14.5 | 3.7 | 25.5 | 1.6 | 9.1 | 5.7 | 27.5 | 8.3 | 36.5 |
Influence of initial protein concentration
The concentration of the protein in the receptor chamber can significantly influence the mass transport. Since Myo is several times smaller than the pore size of M53 concentration dependent Fickian diffusion is expected. The influence of the initial protein concentration on the diffusion profile of Myo through M53 is presented in the ESI Fig. S4.† The concentration of Myo was varied between 0.5 mg L−1, 1.0 mg L−1, and 2.0 mg L−1. The curves show that the cumulative amount of diffusant is directly proportional to the initial concentration of the protein. Thus, the diffusion coefficient Deff for all concentrations is nearly the same (Table 3). For further experiments we chose an initial concentration of 1 mg mL−1 protein in the donor compartment.Size selective separation by diffusion
The diffusion of five model proteins, namely Lys, Myo, Hem, Cat, and Fer through PS-b-P4VP membranes with different pore sizes was investigated. These proteins have been chosen to determine the size selective separation performance of our membranes using proteins with mainly globular shape. The proteins are further characterized e.g. by different isoelectric points (IEP), functional groups, and hydrophilic-to-hydrophobic ratio leading to a variety of possible interactions between the membrane surface area and each protein.Since the membranes are supported by a nonwoven, the impact of the support on the diffusion rate of each model protein was carried out. Diffusion profiles and selectivities of the non-woven are provided in the ESI Fig. S5 and Table S1.† This study proved that the nonwoven itself does not have a significant selectivity towards the proteins. Furthermore the mass transport is very irregular based on the huge mesh sizes of the support and its inhomogeneity. Nevertheless the nonwoven acts as an additional barrier limiting the free diffusion of proteins due to its thickness.
Influence of pH of the release medium
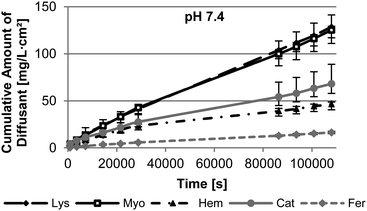
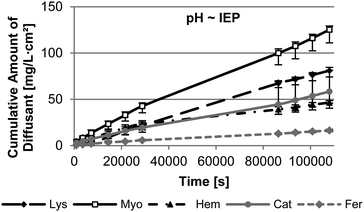
The pH of the release medium is one parameter significantly effecting the interaction and the size of the nanopore and the protein. In order to gain a first insight in the effect of pH, the diffusion through M53 was measured under physiological conditions (pH 7.4) and at pH ∼ IEP of the protein. As depicted in Fig. 5, Lys and Myo show the highest diffusion rate through the membrane M53 attributed to their small hydrodynamic radii. Their mass transport is quite similar at pH 7.4. In general bigger-sized proteins provide a significantly slower diffusion through the membrane. Fer, as the biggest protein, has the lowest diffusion rate. However, under this conditions, Cat diffuses faster through the membrane than Hem. We assume this to be based on the higher hydrophilicity of Cat compared to Hem. Since the pore walls of our membranes are hydrophilic (P4VP), Cat meets less resistance leading to an increased diffusion compared to Hem.The isoelectric point of Myo, Hem, and Fer is almost present under physiological conditions. Therefore the results of the measurement at pH 7.4 can be used. Contrary the IEP of Lys and Cat is found at 11.4 or 5.4, respectively. The diffusion measurements performed at the IEP of these proteins show a decrease in the diffusion rate for both of them (Fig. 6).
This can be attributed to an increased adsorption of the proteins on the membrane, which was carried out in the adsorption studies. This adsorption can induce a lag time in diffusion, as we found for Lys (see ESI Fig S6†). Using a pre-adsorbed membrane, a membrane after adsorption testing, leads to the absence of the lag time. Here the diffusion is nearly comparable to the results at pH 7.4 over the first 8 h of the measurement. However, afterwards the diffusion slows down. Previous studies showed that the diffusion of Lys is slower near to its IEP compared to low pH conditions.12 This is assumed to be based on the missing repulsion of the Lys molecules due to their nearly absent charge at their IEP. Lys exists as monomers or dimers at low pH. On the contrary the protein can assemble to oligomers at a pH close to its IEP.38 Therefore the effective size of the protein is bigger leading to slower diffusion. M53 has a very big pore size compared to the size of Lys of 3.8 nm. Therefore we presume, that the adsorption does not affect the pore size in the initial steps of the measurement of M53 to an extend that an additional resistance for the diffusion of Lys arises.
However, Cat shows a slower diffusion throughout the measurement. We suggest that the high adsorption of Cat at its IEP leads to the reduction of the effective pore size of the membrane and pore blocking. This is supported by SEM analysis of the membrane structure after adsorption testing (Fig. S2†).
Influence of pore size
The diffusion profiles of the five model proteins at physiological pH using the membranes M17, M23, and M34 are depicted in the ESI Fig. S7† and their diffusion coefficients and separation selectivities are listed in Tables 4 and 5. Based on these results we figure out the influence of pore size on the diffusion rate of the proteins and the separation selectivity.| Separation selectivity S | ||||||
|---|---|---|---|---|---|---|
| Protein a/protein b | M17 | M23 | M34 | M53 | PC30 | |
| Lys | Myo | 0.8 | 1.0 | 1.1 | 1.0 | |
| Hem | 3.7 | 4.3 | 3.2 | 3.0 | ||
| Cat | 2.8 | 2.9 | 2.6 | 2.0 | ||
| Fer | 8.7 | 27.3 | 11.5 | 8.3 | ||
| Myo | Hem | 4.7 | 4.3 | 2.8 | 2.9 | |
| Cat | 3.6 | 2.9 | 2.4 | 2.0 | 1.9 | |
| Fer | 11.0 | 27.4 | 10.3 | 8.1 | 6.8 | |
| Hem | Cat | 0.8 | 0.7 | 0.8 | 2.8 | |
| Fer | 2.3 | 6.4 | 3.6 | 2.8 | ||
| Cat | Fer | 3.1 | 9.3 | 4.4 | 4.2 | 3.6 |
The order of the model protein mass transports through M17, M23, and M34 is similar to those of M53.
While a reduction of the pore size from M53 to M34 is in accordance with a decrease of the diffusion constant Deff for the big-sized proteins Cat and Fer, the diffusion of Lys, Myo, and Hem is not affected. The diffusion of proteins in narrow nanopores is effected by a variety of parameters and differs markedly from their free diffusion. A nanopore size of 34 nm to 53 nm is large in scale compared to protein sizes of 3 nm to 7 nm of Lys, Myo, and Hem. As a result the hydrodynamic hindrance for small proteins is not increasing due to interactions between the protein and the pore. This correlates well with our assumption of the mass transport of Lys through M53 at different pH. When pore size and protein size converge, their interaction and electrostatic repulsion increase leading to a decreasing diffusion speed. In the range of pore size this behaviour is found for proteins with outer diameter of 10 nm to 12 nm. These proteins experience a retaining force although the pore size is more than twice as large as the hydrodynamic diameter being in accordance with previous work.16,31 On the one hand the adsorption of proteins reduces the effective pore size as discussed in the analysis of the adsorption measurements. On the other hand electrostatic repulsion, protein structure and process parameters influence the effective size of the protein, the nanopore and their interactions. For M23 a reduction of the diffusion coefficient of all model proteins is found. This effect continues when using M17. In these cases the mass transport is also strongly reduced for small proteins. On the one hand the size of nanopore and small proteins become close leading to increasing interactions and their diffusion slows down. Our results and the restriction of the diffusion of the proteins can be directly linked to the adsorption of the proteins discussed before. On the other hand the surface porosity of the membranes is decreasing from M53 to M17. Therefore the effective diffusion area for the proteins decreases as well, leading to lower mass transport rates.
Table 5 shows the separation selectivities S of the investigated membranes, that are mainly increasing with decreasing pore size of the PS-b-P4VP membrane until it reaches a maximum of 27 for the separation of Myo or Lys and Fer at a pore size of 23 nm. In addition good selectivities for the separation of Hem/Fer and Cat/Fer could be found.
Comparison with commercial track-etched membrane PC30
The performance of our membranes is compared to PC30. The membrane was tested in diffusion experiments using Myo, Cat, and Fer.In comparison PS-b-P4VP membranes with similar pore sizes, M23 and M34, provide significantly higher separation selectivities between Myo/Fer and Cat/Fer. However, their permeability coefficients are lower compared to PC30. This can be attributed to an at least six times larger thickness of PS-b-P4VP membranes compared to PC30. Furthermore an additional resistance arises due to the thickness of the nonwoven support, resulting in a loss of permeability, as already discussed. However, taking into account the thickness of the block copolymer membrane layer the diffusion coefficient Deff of small proteins through PS-b-P4VP membranes is considerably higher than for PC30. This applies to the entire range of pore sizes and can be attributed to the high porosity of our membranes.
Conclusions
In this work we investigated the protein separation performance of self-assembled amphiphilic diblock copolymer membranes. Four PS-b-P4VP membranes varying in pore size from 17 nm to 53 nm were studied concerning clean water fluxes, adsorption and passive diffusion of five proteins. The results were compared with one commercially available polycarbonate track-etched membrane.The water fluxes increase with the pore sizes of the PS-b-P4VP membranes while the flux decline increases as well presumable caused by a higher fouling.
The adsorption of proteins on PS-b-P4VP membranes was influenced by pore size, pH of the media and type of protein. Our results confirm the impact of the interaction of proteins and membranes regarding effective pore sizes and performance. The study provided a detailed basis for both our diffusion experiments and further examination of the membrane characteristics.
For diffusion experiments initial process parameters like stirring speed and the donor concentration were set. Size selective separation by single diffusion of different proteins show that the diffusion coefficient and selectivities for our membranes are significantly higher compared to the polycarbonate membrane. Overall the PS-b-P4VP membrane with pore diameter of 23 nm shows the best theoretical selectivity between myoglobin and ferritin. The difference in diffusion rates indicates a possible effective separation of proteins with only 2 nm difference in size.
Further improvement of the protein separation and membrane flux could be obtained using post-modified PS-b-P4VP membranes. This can be attributed to selective protein pore interactions or decreasing protein adsorption leading to antifouling behaviour. Dual stimuli responsive isoporous membranes as described before37 could enable temperature switchable diffusion of biomolecules and therefore bear a high potential in medical application. The performance of PS-b-P4VP membranes could be improved by reduction of the membrane thickness or increase of the porosity of the substructure.
When it comes to application mixed protein solutions should be studied regarding their adsorption and diffusion behaviour as well.
Acknowledgements
The authors thank Brigitte Lademann for the synthesis of the polymers, Kristian Buhr for the preparation of the membranes, Sofia Rangou for scientific discussion, Barbara Bajer for the help with the adsorption and diffusion measurements, Maren Brinkmann for the GPC measurements, Silvio Neumann for the NMR and Jan Pohlmann for construction of the automatic water flux set-up. This work is financially supported by FP7 EU-project SELFMEM under grant agreement NMP3-SL-2009-228652.Notes and references
- A. Saxena, B. P. Tripathi, M. Kumar and V. K. Shahi, Adv. Colloid Interface Sci., 2009, 145, 1–22 CrossRef CAS PubMed.
- R. van Reis and A. Zydney, Curr. Opin. Biotechnol., 2001, 12, 208–211 CrossRef CAS.
- G. Jeon, S. Y. Yang and J. K. Kim, J. Mater. Chem., 2012, 22, 14814 RSC.
- G. M. Geise, H.-S. Lee, D. J. Miller, B. D. Freeman, J. E. McGrath and D. R. Paul, J. Polym. Sci., Part B: Polym. Phys., 2010, 48, 1685–1718 CrossRef CAS.
- N. Savage and M. Diallo, J. Nanopart. Res., 2005, 7, 331–342 CrossRef CAS.
- B. Van Der Bruggen, C. Vandecasteele, T. Van Gestel, W. Doyen and R. Leysen, Environ. Prog., 2003, 22, 46–56 CrossRef CAS.
- S. Y. Yang, I. Ryu, H. Y. Kim, J. K. Kim, S. K. Jang and T. P. Russell, Adv. Mater., 2006, 18, 709–712 CrossRef CAS.
- B. B. Lakshmi and C. R. Martin, Nature, 1997, 388, 758–760 CrossRef CAS PubMed.
- P. Stroeve and N. Ileri, Trends Biotechnol., 2011, 29, 259–266 CrossRef CAS PubMed.
- W. M. Clark, A. Bansal, M. Sontakke and Y. H. Ma, J. Membr. Sci., 1991, 55, 21–38 CrossRef CAS.
- W. S. Dai and T. A. Barbari, Biomaterials, 2000, 21, 1363–1371 CrossRef CAS.
- C. Mattisson, P. Roger, B. Jönsson, A. Axelsson and G. Zacchi, J. Chromatogr. B: Biomed. Sci. Appl., 2000, 743, 151–167 CrossRef CAS.
- B. C. Robertson and A. L. Zydney, J. Colloid Interface Sci., 1990, 134, 563–575 CrossRef CAS.
- P. van Rijn, M. Tutus, C. Kathrein, L. Zhu, M. Wessling, U. Schwaneberg and A. Boker, Chem. Soc. Rev., 2013, 42, 6578–6592 RSC.
- N. Ileri, R. Faller, A. Palazoglu, S. E. Letant, J. W. Tringe and P. Stroeve, Phys. Chem. Chem. Phys., 2013, 15, 965–971 RSC.
- C. C. Striemer, T. R. Gaborski, J. L. McGrath and P. M. Fauchet, Nature, 2007, 445, 749–753 CrossRef CAS PubMed.
- S. Y. Yang, J. A. Yang, E. S. Kim, G. Jeon, E. J. Oh, K. Y. Choi, S. K. Hahn and J. K. Kim, ACS Nano, 2010, 4, 3817–3822 CrossRef CAS PubMed.
- K. V. Peinemann, V. Abetz and P. F. Simon, Nat. Mater., 2007, 6, 992–996 CrossRef CAS PubMed.
- R. M. Dorin, W. A. Phillip, H. Sai, J. Werner, M. Elimelech and U. Wiesner, Polymer, 2014, 1, 347–353 CrossRef PubMed.
- J. Hahn, V. Filiz, S. Rangou, J. Clodt, A. Jung, K. Buhr, C. Abetz and V. Abetz, J. Polym. Sci., Part B: Polym. Phys., 2013, 51, 281–290 CrossRef CAS.
- J. Hahn, V. Filiz, S. Rangou, B. Lademann, K. Buhr, J. I. Clodt, A. Jung, C. Abetz and V. Abetz, Macromol. Mater. Eng., 2013, 298, 1315–1321 CrossRef CAS.
- A. Jung, V. Filiz, S. Rangou, K. Buhr, P. Merten, J. Hahn, J. Clodt, C. Abetz and V. Abetz, Macromol. Rapid Commun., 2013, 34, 610–615 CrossRef CAS PubMed.
- A. Jung, S. Rangou, C. Abetz, V. Filiz and V. Abetz, Macromol. Mater. Eng., 2012, 297, 790–798 CrossRef CAS.
- M. Gallei, S. Rangou, V. Filiz, K. Buhr, S. Bolmer, C. Abetz and V. Abetz, Macromol. Chem. Phys., 2013, 214, 1037–1046 CrossRef CAS.
- J. I. Clodt, S. Rangou, A. Schroder, K. Buhr, J. Hahn, A. Jung, V. Filiz and V. Abetz, Macromol. Rapid Commun., 2013, 34, 190–194 CrossRef CAS PubMed.
- P. Madhavan, K. V. Peinemann and S. P. Nunes, ACS Appl. Mater. Interfaces, 2013, 5, 7152–7159 CAS.
- L. Oss-Ronen, J. Schmidt, V. Abetz, A. Radulescu, Y. Cohen and Y. Talmon, Macromolecules, 2012, 45, 9631–9642 CrossRef CAS.
- D. S. Marques, U. Vainio, N. M. Chaparro, V. M. Calo, A. R. Bezahd, J. W. Pitera, K.-V. Peinemann and S. P. Nunes, Soft Matter, 2013, 9, 5557 RSC.
- R. M. Dorin, D. S. Marques, H. Sai, U. Vainio, W. A. Phillip, K.-V. Peinemann, S. P. Nunes and U. Wiesner, ACS Macro Lett., 2012, 1, 614–617 CrossRef CAS.
- S. Rangou, K. Buhr, V. Filiz, J. I. Clodt, B. Lademann, J. Hahn, A. Jung and V. Abetz, J. Membr. Sci., 2014, 451, 266–275 CrossRef CAS PubMed.
- X. Qiu, H. Yu, M. Karunakaran, N. Pradeep, S. P. Nunes and K. V. Peinemann, ACS Nano, 2013, 7, 768–776 CrossRef CAS PubMed.
- A. Jayalakshmi, S. Rajesh and D. Mohan, Appl. Surf. Sci., 2012, 258, 9770–9781 CrossRef CAS PubMed.
- B. Steffansen, B. Brodin and C. U. Nielsen, in Molecular Biopharmaceutics: Aspects of Drug Characterisation, Drug Delivery and Dosage Form Evaluation, ed. B. Steffansen, B. Brodin and C. U. Nielsen, Pharmaceutical Press, London, Chicago, 1st edn, 2009, ch. 3.2, pp. 135–152 Search PubMed.
- C. H. Lu and W. J. Lin, J. Biomed. Mater. Res., 2002, 63, 220–225 CrossRef CAS PubMed.
- Y. S. Polyakov and A. L. Zydney, J. Membr. Sci., 2013, 434, 106–120 CrossRef CAS PubMed.
- S. Yamane, K. Takayama and T. Nagai, J. Controlled Release, 1998, 50, 103–109 CrossRef CAS.
- J. I. Clodt, V. Filiz, S. Rangou, K. Buhr, C. Abetz, D. Hoche, J. Hahn, A. Jung and V. Abetz, Adv. Funct. Mater., 2013, 23, 731–738 CrossRef CAS.
- F. Carlsson, M. Malmsten and P. Linse, J. Phys. Chem. B, 2001, 105, 12189–12195 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ra47306f |
| This journal is © The Royal Society of Chemistry 2014 |