Hybrid assembly of DNA-coated gold nanoparticles with water soluble conjugated polymers for studying protein–DNA interaction and ligand inhibition†
Steven Lukmana,
Khin Moh Moh Aunga,
Michelle Gek Liang Limb,
Shuzhen Hongb,
Si Kee Tanb,
Edwin Cheung*bcd and
Xiaodi Su*a
aInstitute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), 3 Research Link, Singapore 117602. E-mail: xd-su@imre.a-star.edu.sg
bCancer Biology and Pharmacology, Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), 60 Biopolois Street, Singapore 138672. E-mail: cheungcwe@gis.a-star.edu.sg
cDepartment of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore
dSchool of Biological Sciences, Nanyang Technological University, Singapore
First published on 14th January 2014
Abstract
Metal nanoparticles (mNPs) have unique optical properties arising from the localized surface plasmon resonance. They have been extensively used as colorimetric probes for chemical and biological analysis, exploiting particle aggregation-induced color change. They can also support fluorimetric detection based on Förster resonance energy transfer (FRET) or nanoparticle surface energy transfer (NSET) with proximal fluorophores. In this paper, luminescent water soluble conjugated polymers (CPs) and dsDNA-coated gold NPs (AuNPs) are used as collaborative sensing elements for studying protein–dsDNA interactions. The hybrid materials-based assays exploit the phenomena of (1) CPs' fluorescence emission can be quenched by AuNPs due to CPs–DNA interactions on AuNPs surface and (2) protein binding to DNA can change the surface charge of the dsDNA–AuNPs conjugates that in turn change the degree of CPs quenching. Three CPs of bearing with different charge and emitting at different wavelength are used to construct the hybrid sensors for determining protein–DNA interactions in terms of sequence selectivity, binding affinity and binding stoichiometry. Depending on the initial quenching, determined by CPs' charge property and emission wavelength relative to the absorption peak of 13 nm AuNPs, “light-on”, “light-off”, and “two way” assays have been constructed that are capable of studying proteins of known or unknown charge properties. We have demonstrated the concept for two important oncogenic factors, i.e. FoxA1 (forkhead boxA1) and AP-2γ (activating enhancer binding protein 2 gamma). They are pivotal in regulating the transcriptional activity of estrogen receptor alpha and controlling the expression of estrogen-responsive breast cancer cells. Determination of their DNA binding properties can reveal how they regulate the transcriptional activity of estrogen receptor. The hybrid sensors have been extended to screen small molecular weight ligands that can inhibit FoxA1– and AP-2γ–DNA complex formation. Identification of inhibitors as drug candidates targeting these two transcription factors could be an alternative in treating breast cancer, in particular those that have become endocrine resistant.
Introduction
Gold nanoparticles (AuNPs) have unique optical properties arising from their ability to support localized surface plasmon resonance (LSPR).1–3 AuNPs and other metal nanoparticles can support colorimetric bioaffinity assays exploiting interparticle distance-determined solution color change (under bright field or dark field measurement)1–7 and fluorimetric assays due to their super quenching to approximate fluorophores.5–7 With the colorimetric principle, bioaffinity binding events (i.e. DNA–DNA, DNA–metal ions, DNA–DNA binding drugs, and protein–DNA, peptide–sugar, etc.) and biological processes (i.e. protein phosphorylation, DNA cleavage etc.) can be detected due to their control of NPs' dispersion and aggregation status detected as specific solution color. For studying protein–DNA interactions, for example, a few assay designs have been reported using either unmodified gold nanoparticles (AuNPs)8,9 or DNA-conjugated AuNPs.10–12 In the unmodified AuNPs-based assays for transcription factors (TFs) and single stranded DNA binding protein (SSB), protein–DNA complexes (i.e. TFs with dsDNA and SSB with ssDNA) can be detected based on their superior protection of citrate ion-coated AuNPs against salt-induced aggregation, relative to protein and DNA without forming stable complex.8,9 In the assays involving DNA-conjugated mNPs, one example is to use two sets of double stranded DNA (dsDNA) modified mNPs, each of the DNA carrying a half site or a segment of a functional DNA sequence for the TF of interest.10 The detection of specific protein–DNA binding is founded on the premise that the mixture of the two sets of dsDNA–mNPs experiences a remarkable particle aggregation, whereas the aggregation can be retarded in the presence of specific protein that binds and introduces steric protection forces between particles. Alternatively, Ou et al. utilized two sets of single stranded DNA (ssDNA)-conjugated DNA to form crosslinked AuNPs network through interparticle DNA hybridization.11 Protein binding to the preformed dsDNA network linkers protects the DNA linkers from cleavage by exonuclease III (Exo III), which in turn protect the blue AuNPs aggregates. In the case when the protein to be studied carries two binding sites to a DNA element, protein binding can be detected by its crosslinking of DNA-conjugated AuNPs, without relying on salt screening and enzymatic DNA cleavage.12 The above mentioned methods are all based on particle aggregation/dispersion related color change for detecting protein–DNA complex formation and sequence specificity. However, they encounter limitations of (1) nonspecific aggregation of bare particles, (2) inadequate sensitivity to differentiate subtle affinity difference induced by single nucleotide variation, (3) slow response in relation to the cleavage of DNA linkers inside the crosslinked AuNPs network, or (4) extensive optimization of enzymatic reaction conditions (enzymatic reaction is very sensitive to temperature and ionic strength of the liquid medium).Water-soluble conjugated polymers (CPs) are promising materials for biosensing and imaging due to their unique electrical and optical properties.13,14 Coupling CPs' excellent photon harvesting property and charge property with high extinction coefficient AuNPs has led to the development of fluorimetric chemical nose/tongue for differentiating specific proteins, pathogen or cells through AuNPs mediated by Förster resonance energy transfer (FRET) or nanomaterial surface energy transfer (NSET).15–17 in one example, a handful of cationic AuNPs and anionic CPs (ACPs) form electrostatically complementary complexes that results in fluorescence quenching of the polymers. Addition of negatively charged analytes (proteins or cells) disrupts the AuNP–ACPs complexes, resulting in fluorescent recovery. Similar principle has been reported for detecting cysteine, according to the strong affinity of cysteine to gold that can displace the adsorbed ACPs, leading to fluorescence recovery.18 To better understand the fluorescent quenching between metal NPs and CPs, Xu et al. has conducted a series studies with negatively charged gold and silver nanoparticles of different sizes to establish size-dependent quenching efficiency using cationic CPs (CCPs).19–21 Besides the fluorometric assays, CPs–AuNPs combination have been used for colorimetric assay of ssDNA and ssDNA binding molecules because a CCP can bind to ssDNA and specifically sequesters ssDNA for stabilizing AuNPs.22
In this study we further harness the excellent photon harvesting and charge properties of water soluble CPs and the super quenching property of AuNPs to develop a simple, rapid, and sensitive methodology for studying transcription factor (TF)–DNA interactions and for screening ligand inhibition that can facilitate drug discovery research. Three water soluble CPs of different emission wavelength and charge properties (ACP and CCP) have been chosen to couple with double-stranded DNA conjugated AuNPs that can ascertain the energy/electron transfer due to the broad absorption spectra of AuNPs. With two oncogenic TFs, i.e. FoxA1 (forkhead boxA1) and AP-2γ (activating enhancer binding protein 2 gamma), and 9 compounds from NIC library, we have demonstrated the concept and proved the generality. Depending on the choice of CPs in terms charge and emission wavelength relative to the absorption wavelength of AuNPs (520 nm), the assay can be constituted into “light-on”, “light-off”, or “two-way” models for detecting sequence dependent protein–DNA binding for proteins of known or unknown charge properties, quantifying affinity constant, and determine the stoichiometry.
Establishing sequence specific protein–DNA interactions characteristics and ligand inhibition are important in biomedical research. FoxA1 and AP-2γ are TFs in the estrogen signalling pathway that functions as pioneer factors in determining the binding, chromatin-looping, and gene transcription-mediated by ERα.23,24 A detailed characterization of the DNA binding properties of FoxA1 and AP-2γ is therefore of great importance to understand how these factors regulate the transcriptional activity of estrogen receptor. Identification of small ligand inhibitor for these two TFs presents alternative for breast cancer therapy.
Results and discussion
Characterization of dsDNA–AuNPs conjugate and initial interaction (quenching) of CPs with (by) dsDNA–AuNPs conjugates
Double stranded DNA (dsDNA) conjugated AuNPs is one of the key sensing elements in this study (DNA sequences used on this study are in Table 1). We have first characterized the dsDNA–AuNPs conjugates. Fig. 1 shows the UV-vis spectrum of AuNPs and AuNPs conjugated with the 17 mer DNA (Probe 2) before and after FoxA1 binding. The successful attachment of the DNA to AuNPs can be confirmed by the slight red shift of the peak wavelength of 4 nm. The surface charge and hydrodynamic size of the 13 mer AuNPs before and after DNA conjugation, as well as the surface coverage of the dsDNA on AuNPs are characterized (Table 2). The hydrodynamic size expansion from 20.4 ± 2.18 nm to 31.9 ± 0.91 nm (i.e. an increase of 11.5 nm) confirmed that the 17 bp dsDNAs are fully extended like a rigid rod on the surface of AuNPs, based on the fact that every 10 bp of a DNA double helix is approximately 3.4 nm in length25 (please note, in this estimation the C6 thiol linker is not taken into consideration). From zeta potential measurements, after DNA conjugation the surface charge density is slightly reduced from −39.2 ± 3.5 mV to −32.7 ± 2.8 mV, but is still substantial to act as molecular screen that wanes the negative charge. Putting together the peak wavelength shift, expanded hydrodynamic size, and the change of surface charge, we have confirmed the successful conjugation of the dsDNA on AuNPs.| Name | Sequence |
|---|---|
| Probe 1 (for FoxA1) | 5′-CACTTT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CAAAGC-3′ CAAAGC-3′ |
| Probe 1-comp | 5′-GCTTTGCAAACAAAGTG-3′ |
| Probe 2 (for FoxA1) | 5′-GTACTGTAAATAAAACT-3′ |
| Probe 2-comp | 5′-AGTTTTATTTACAGTAC-3′ |
| Probe 3 (for FoxA1) | 5′-![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TAAATA TAAATA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) -3′ -3′ |
| Probe 3-comp | 5′-CTGCACTATTTACTTGGCA-3′ |
| wtR3 (for AP-2γ) | 5′-AAAGTGCCCAGAGCCCATG-3′ |
| wtR3-comp | 5′-CATGGGCTCTGGGCACTTT-3′ |
| mtR3 (for AP-2γ) | 5′-AAAGT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CAGA CAGA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CATG-3′ CATG-3′ |
| mtR3-comp | 5′-CATGGATTCTGAATACTTT-3′ |
 | ||
| Fig. 1 UV-vis spectrum of AuNPs and AuNPs coated with 17 mer dsDNA (Probe 2), before and after FoxA1 binding. | ||
| Particles | Hydrodynamic diametera (nm) | Zeta potential (mV) | DNA surface coverageb (molar ratio) |
|---|---|---|---|
| a Measured by dynamic light scattering.b Molar ratio of dsDNAs that bound to the surface of AuNPs measured with thiazole orange (detail in ESI).c FoxA1 to dsDNA ratio is 5. | |||
| Bare AuNPs | 20.4 ± 2.18 | −39.2 ± 3.5 | N.A. |
| dsDNA–AuNPs | 31.9 ± 0.91 | −32.7 ± 2.8 | 99.7 ± 6.0 |
| FoxA1 bound dsDNA–AuNPsc | 52.8 ± 1.7 | −25.8 ± 2.5 | |
| ACP-430 bound FoxA1–dsDNA–AuNPs | 53.4 ± 1.9 | −26.3 ± 2.3 | |
| CCP-410 bound FoxA1–dsDNA–AuNPs | 53.8 ± 0.3 | −24.6 ± 5.7 | |
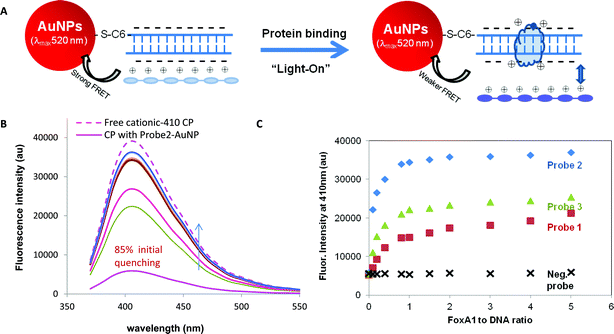
To study the ability of dsDNA–AuNPs conjugates to quench the CPs emission, nanoparticles and CPs were mixed at equal molar; CPs fluorescence emission was measured and compared with the same concentration of CPs without the nanoparticles. Fig. 2B shows the emission spectra of the three CPs with and without addition of the 17 bp dsDNA–AuNPs (or bare AuNPs). The absorbance spectra of AuNPs (or dsDNA–AuNPs) is given as a reference to show the overlap integral of CPs (donors) emission and AuNPs (acceptor) absorption spectra that would dictate the initial quenching of CPs by dsDNA–AuNP. All CPs show detectable drop in fluorescent intensity due to the interaction with the dsDNA surrounding AuNPs that brings the polymer close to AuNPs for energy and/or electron transfer and thus non-radiative decay. The degree of quenching is largely determined by the charge of the polymer and the spectrum overlap with the AuNPs absorption. For example, the ACP-430 (λem = 430 nm) showed slight quenching of ∼15% by the dsDNA–AuNPs. The emission peak was slightly blue shifted from 430 nm to 420 nm. The slight increase in energy gap of this polymer could be due to the change in conformation upon interacting with dsDNA. Since both ACP-430 polymer chain and dsDNA possess net negative charges, electrostatic attraction could not exist between them and in fact there should be remarkable repulsion. Thus the slight quenching could presumably due to the hydrophobic interaction between the polymer's non-polar carbon chains with the molecular backbone of hydrophobic DNA that preferred to stay away from polar water molecules in solvent. This speculation was proven by decreasing the polarity of the solvent (by adding chloroform). As the chloroform content was increased (up to 40 vol%), the degree of fluorescence quenching diminishes gradually up to 40 vol% chloroform, in which only small quenching could be detected (ESI, Fig. S1†). The reasoning behind this observation is CPs could minimize its free energy, increase the entropy by having more random configuration, in the less polar organic solvent without have to interact with the dsDNA. To confirm no appreciable energy or electron transfer occurred in the absence of AuNPs, CPs were incubated individually with only free dsDNA or protein. Neither dsDNA nor protein quenched the fluorescence of CPs (ESI, Fig. S2†). The change in fluorescence intensity was only observed with the dsDNA coated AuNPs. For the other negatively charged polymer, ACP-560 (λem = 560 nm), the overlap integral with the AuNPs absorption (λmax = 520 nm) is more than ACP-430 (λem = 430 nm). Despite the same weak interaction with dsDNA, this ACP is quenched at a larger degree (∼50%) than the ACP-430. This indicates that larger degree of overlap between CP's emission and AuNPs' extinction spectrum is essential for strong energy transfer and thus more fluorescence quenching.
In comparison with the negatively charged ACPs, positively charged CCP-410 (λem = 410 nm) has pretty similar overlapping spectra as ACP-430. However, a much stronger quenching (∼85%) was observed. This is attributable to the positively intrinsic charge of the polymer chain that presents electrostatic interaction with dsDNA–AuNPs. Both ACP-560 and CCP-410 does not exhibit any shift in the emission peak. One possible explanation is both these CPs has higher hydrophobicity and thus less likely to change conformation upon interacting with dsDNA, unlike the ACP-430 which is more hydrophilic. The stronger hydrophilic characteristic of ACP-430 is evinced as higher solubility in water.
To understand the effect of the dsDNA coating on the CP quenching by AuNPs, we compared the quenching behaviour of bare AuNPs and dsDNA–AuNPs (Fig. 2B, dotted lines versus dashed lines). As expected, bare AuNPs quenched the CPs stronger than the dsDNA–AuNPs conjugates. Particularly, the ACP-430 and ACP-560 were quenched roughly by 20% and 65%, compared to 15% and 50% quenching exerted by dsDNA–AuNPs; and the CCP-410 was almost completely quenched by the unmodified AuNPs. The enhanced quenching by bare AuNPs can be reasoned as that CPs could approach even closer to bare AuNP surface by direct attachment, and the AuNPs–fluorophore quenching is a distance dependent process (the closer the more efficient). The dsDNA coating (∼11.5 nm thick) on the conjugates acted as spatial hindrance that prevents CPs from direct attachment to AuNP, and thus depletes the quenching efficiency.
Detecting FoxA1–DNA interaction using ACP-430 by enhanced fluorescence quenching, i.e. a “light-off” assay
In Fig. 1 we have shown that FoxA1 binding to Probe 2-coated AuNPs gives only minor peak wavelength shift (∼4 nm). This minor peak shift is far less convincing to measure the successful protein binding and leaves no room to study DNA of weaker affinity (Probe 2 is of the highest affinity to FoxA1 among other DNA to be detected later on). It is then essential to introduce other sensing strategies. Remarkable quenching of CPs by dsDNA–AuNPs has been observed in the previous session. It can be exploited to “amplify” the protein binding under the fluorescence measurement.Using ACP-430 as an example, we first demonstrated a “light-off” assay to test the binding of FoxA1 to Probe 1, Probe 2, and Probe 3, using respective DNA-conjugated AuNPs. The principle is presented schematically in Fig. 3A. This anionic polymer only exhibits a weak interaction with the dsDNA–AuNPs, detected as a smaller degree of initial quenching (15%). FoxA1 has isoelectric point (pI) of ∼8.9,26 hence it is positively charge at pH 7.4. The binding of this protein to dsDNA–AuNPs has been further confirmed by the remarkably increase hydrodynamic size and the reduced negative charge of the nanoparticles (Table 2). Due to the binding of the positively charged FoxA1, the fluorescence intensity of ACP-430 was further suppressed as shown for the Probe 2 DNA in Fig. 3B. The fluorescence intensity decreased with increasing concentration of FoxA1 before reaching a saturation value for all three tested probes (Fig. 3C). We reasoned that the binding of FoxA1 to the DNA neutralized the negative charge of dsDNA–AuNP to certain degree as evidenced by the zeta potential data in Table 2 (from −32.7 ± 2.8 to −25.8 ± 2.5 mV), and thus more anionic polymer chains can access to the AuNPs due to reduced charge repulsion. When the CPs were close enough to the surface, energy/electron transfer take place and act as radiationless pathway for the excited electrons to decay to ground state, and thus the quenching. The further increase of negative charge density from FoxA1-bound dsDNA–AuNPs to −26.3 ± 2.3 is supportive of the adsorption of negatively charged ACP-430 chains to FoxA1-bound dsDNA–AuNPs (Table 2). When Probe 1 and Probe 3 were tested, we found that the prominent significance was each dsDNA–AuNP responded differently with the protein and thereby the measured fluorescence intensities for the same concentration of protein used were different, following an order of Probe 2 > Probe 3 > Probe 1 in their quenching strength (Fig. 3C). A negative control was also measured, which was mixture of mtR3–AuNP and FoxA1. The fluorescent intensity did not change with addition of FoxA1. This trend was observed for any concentration of protein added, though the differentiation became more vivid at higher concentration.
Detecting FoxA1–DNA interactions using CCP-410 by fluorescence restoration, i.e. a “light-on” assay
While the above hybrid sensor using ACP-430 with limited initial quenching can detect protein–DNA binding through enhanced fluorescent quenching, the “light-off” model offered a relatively low sensitivity because of the weaker interaction between ACP and dsDNA, which translates into smaller change in fluorescence intensity (∼30% of further reduction) upon protein binding. Differing from the ACP-430, mixing the positively charged CCP-410 with dsDNA–AuNP led to a drastic initial quenching of 85% (Fig. 2B) of the fluorescence intensity. As discussed earlier, the drastic initial quenching can be attributed to the electrostatic attraction interaction between the polymer chains and the dsDNA that can bring sufficient amount of polymer chains close to the AuNPs. When this polymer is used to constitute the hybrid sensor, protein binding can be detected as fluorescent recovery (or light-on) (Fig. 4A). When FoxA1 was added to interact with the dsDNA–AuNPs, followed by addition of CCP-410, the fluorescence intensity gradually recovered with increasing FoxA1 concentration (Fig. 4B). As discussed earlier and proven by the zeta potential value (Table 2), we reasoned that when FoxA1 (positively charged at pH 7.4) binds to the DNA, the overall negative charge of the dsDNA–AuNPs complex is neutralized that weakens the electrostatic binding between the positively charged CP chains and the dsDNA–AuNPs, which is detected as reduced fluorescence quenching or fluorescent restoration. At the highest FoxA1 concentration (ratio 5 to dsDNA) the CCP-410 binding is minimal as shown by almost completely depleted quenching in Fig. 4 and the insignificant change of the zeta potential of the FoxA1-bound AuNPs (Table 2). For the three DNA probes tested for FoxA1, the extent of fluorescence recovery followed closely the same order among the three probes: Probe 2 > Probe 3 > Probe 1 (Fig. 4C). Similar control experiment was conducted by a negative control DNA (mtR3–AuNP in this case), which did not give appreciable change in fluorescent intensity. Interestingly, the high degree of initial quenching offers a “light-on” assay that is more appealing since it is suitable for visual detection (inset in Fig. 4B).Two-way detecting of FoxA1–DNA interactions using ACP-560
Using an ACP and a CCP of either limited initial quenching or drastic initial quenching (i.e. ACP-430 and CCP-410), we have shown that we can detect positively charged FoxA1 by either enhanced fluorescent quenching (when the initial quenching in the absence of protein is low) or reduced quenching (when the initial quenching in the absence of protein is high). In those examples, the positively charged protein can be detected by its role of either attracting negatively charged CPs (enhanced quenching) or repealing positively charged CP (reduced quenching). Here we show that the ACP that emits near AuNPs absorption peak of 520 nm (ACP-560) allows for “two-way” protein detection for both positive and negative proteins. The modest initial quenching of 50% (Fig. 2B) due to negative charge and the closer spectrum overlay with the 13 nm AuNPs provides room for both “light-on” and “light-off” detection (Fig. 5A). This negatively charged polymer is unlike the other negatively charged polymer (ACP-430), where the slight initial quenching for ACP-430 gave it disadvantage to be used for detecting negatively charged protein, because the range of the limited fluorescence restore can be explored.To test the dual applicability of this ACP-560, FoxA1 was measured at two conditions, namely pH 7.4 and pH ∼ 9.5, which turns the protein either positively or negatively charged, respectively. At pH 7.4, FoxA1 (positively charged) is detected by ACP-560 similarly as ACP-430, in which addition of FoxA1 generated gradual increase in quenching (Fig. 5A and B). At pH 9.5, however, FoxA1 (negatively charged) binding caused fluorescence recovery (Fig. 5C), following similar trend as the fluorescence recovery process when FoxA1 binding was probed with CCP-410 at pH 7.4. The magnitude of change in intensity decreased from Probe 2 > Probe 3 > Probe 1. This type of CP/DNA–AuNPs assembly is particularly suitable to study protein when the isoelectric point is not known, which is not often possible by other FRET sensors involving CP and organic dye donor and acceptor pair.
Quantitative determination of binding constant (Kd) and stoichiometry (n)
Qualitatively, the affinity of FoxA1 to different DNA probes could be identified with the degree of change in fluorescence intensity. For quantification purpose, binding constant was calculated from FoxA1 titration data. The extent of CP quenching merely by dsDNA–AuNP defined the baseline for the subsequent calculation of binding constant and stoichiometry. With the ACP-410/AuNPs sensor as example, with the [protein] dependent fluorescence intensity scales, protein–DNA Kd, is calculated through eqn (1):| (F0 – F)/(F – Fsat) = ([protein]/Kd)n | (1) |
| Method | FoxA1 | AP-2γ | |||
|---|---|---|---|---|---|
| Probe 1 | Probe 2 | Probe 3 | wtR3 | mtR3 | |
| FA | 230.5 ± 34.1 | 16.0 ± 3.6 | 28.9 ± 6.5 | 50.6 ± 14.2 | 293.2 ± 11.1 |
| ACP-430/DNA–AuNPs hybrid sensor | 31.3 ± 4.4 | 4.5 ± 0.2 | 12.5 ± 1.1 | 12.4 ± 0.7 | 53.1 ± 4.0 |
| ACP-560/DNA–AuNPs hybrid sensor | 32.9 ± 4.0 | 5.8 ± 0.8 | 14.8 ± 1.5 | 14.6 ± 0.5 | 54.7 ± 1.3 |
| CCP-410/DNA–AuNPs hybrid sensor | 34.1 ± 3.5 | 5.0 ± 0.7 | 12.4 ± 0.4 | 13.0 ± 1.4 | 50.4 ± 3.2 |
With the hybrid sensor and the conventional FA biochemistry method, we have collectively confirmed that Probe 2 (perfect sequence) has the highest binding affinity to FoxA1, followed by Probe 3 (mutation at the flanking region) and Probe 1 (mutated at the centre part). Importantly, although all methods show the same affinity trend for FoxA1 binding, the absolute Kd values are different, with those from our hybrid sensors being relatively lower but closer to those from FA. Such differences are common and expected since different measurement methods rely on different principles. The similar values from our hybrid sensors and FA assay can be attributed to the similar homogenous phase complex formation assay method.
According to the Kd value measured by the current method and FA, FoxA1 can bind to FoxA1 site (Probe 2 and Probe 3) efficiently (with higher affinity) when the core consensus sequence C/AAAC/T is preserved. Changing the flanking sequence (in Probe 3) only leads to a slight decrease in affinity. On the other hand, the affinity drops significantly for Probe 1 that contains ‘T’ to ‘C’ variant in the core sequence. Dong et al. also reached the same conclusion, who studied the effect of polymorphism in FoxA1 binding site.27 Current experimental setup can differentiate change in FoxA1 binding affinity by subtly altering the FoxA1 binding consensus. Carroll et al.23 suggested that many of the single nucleotide polymorphisms associated with breast cancer risk may be modulating biological process, namely FoxA1 binding, a key regulator of ERα activity. Therefore, this hybrid assembly can be potentially employed to determine the effect of modification of risk locus on FoxA1 function and ERα-dependent transcription. Furthermore, the slope of the logarithmic plot gave n ∼ 1, suggesting that FoxA1 bound to DNA in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. This capability of measuring binding stoichiometry is of another advantage of the hybrid sensor.28
1 ratio. This capability of measuring binding stoichiometry is of another advantage of the hybrid sensor.28
Generality of the assay demonstrated with AP-2γ and its DNA elements
The hybrid sensors were utilized to study a second TF, AP-2γ (pI ∼ 7.8)29 to test the generality of the method. Both ACP-430 and CCP-410 and were used to test the protein binding at pH 7.4 (Fig. S4†). Similar to FoxA1, Ap-2γ binding is detected as either the further quenching to ACP-430 or fluorescence recovery for CCP-410 (ESI, Fig. S4†). From the relative decrease and increase in fluorescent intensity of CP with increasing concentration of AP-2γ (ESI, Fig. S5A and B†), Kd and n were determined (Fig. S5C†). The calculated Kd (given in Table 4) indicate that AP-2γ bound stronger to wtR3 than to mtR3. The wtR3 probe contains palindromic sequences, 5′-GCCN3GCC-3′, which appears to be the core recognition elements for AP-2 binding site.29 When the base was mutated to their purine and pyrimidine counterparts, AP-2γ binding affinity was affected drastically.| Ligand | Name | Mw | Chemical formula |
|---|---|---|---|
| 1 | Quinobene | 931 | C32H22N6O14S4·4Na |
| 2 | Dawson | 3015 | K6P2Mo18O62 |
| 3 | [[(Z)-(3-Oxopyridin-2-ylidene)methyl]amino]thiourea (picolinaldehyde) | 196 | C7H8N4OS |
| 4 | 8-Amino-10-phenylphenazin-2-one | 287 | C18H13N3O |
| 5 | 2-Bromo-1H-phenalen-1-one | 259 | C13H7BrO |
| 6 | Lomofungin | 314 | C15H10N2O6 |
| 7 | N,N-Dimethyldaunorubicin hydrochloride | 592 | C29H33NO10·ClH |
| 8 | 6H-Imidazo[4,5,1-de]acridin-6-one, 5-[2-(diethylamino) ethylamino]-8-methoxy-1-methyl-, dihydrochloride | 451 | C22H26N4O2·2ClH |
| 9 | Quinacrine hydrochloride/mepacrine | 473 | C23H30ClN3O·2ClH |
The finding that AP-2γ binds to a palindromic recognition sequence implies that AP-2γ may interact with DNA as a multimer. Moreover, the region of AP-2γ required for DNA binding is quite large and suggests that, in addition to direct DNA contact, protein–protein contact between AP-2γ molecules may help to stabilize the protein–DNA interaction further. From our current assay, the slope of the plot was ∼2, which confirms that AP-2γ binds to DNA as dimer. This result is in agreement with the previous finding for AP-2γ, which was shown to exist as stable dimmers in solution and would lose its ability to bind DNA if the proteins does not dimerize.30
Screening ligand inhibition to protein–DNA interactions
Some organic molecules can bind to proteins and change their DNA binding properties. Identification and characterization of small organic molecules that inhibit or weaken protein binding to DNA may lead to the discovery of new therapeutic drugs. In this study, 9 ligands selected from NCI compound library (Table 4) were tested for their effect on FoxA1– and AP-2γ–DNA binding. Fig. 6 shows examples for FoxA1 and a few ligands detected by ACP-430/Probe 2–AuNPs and CCP-410/Probe 2–AuNPs hybrid sensors, with the negative control DNA–AuNPs as reference. With both polymers, ligands 1 and 2 are found substantially inhibiting FoxA1–DNA binding. The respective CP emission is very similar to the cases where no protein is added. Other ligands, i.e. ligands 3 and 4, show negligible effect. The polymer emission is close to the case where FoxA1 is added without any ligands. Ligand 6 is found to inhibit FoxA1 binding to a partial degree. The same partial inhibition is observed for ligands 7 and 8, enervates FoxA1 binding affinity but not as effective as ligands 1 and 2. Fig. 7 shows the ligand inhibition tests for all 9 ligands by the ACP-430/Probe 2–AuNPs hybrid sensor and the traditional FA that relies on fluorescent labeled DNA. In the fluorescence anisotropy measurement, when FoxA1 binds to FAM-labeled Probe 2, the overall size of the molecule increases. This will hamper the capability of FAM-Probe 2 to rotate freely. The sluggish motion is seen as increase in fluorescence anisotropy of the FoxA1–Probe 2 entity against the free Probe 2. Successful ligand inhibition (by ligands 1 and 2 for example) is detected as lower fluorescence anisotropy value, vice versa. For the 9 ligands tested, the FA result agrees well with the outcome concluded from current hybrid sensor assay. In the presence of ligands 3, 4, 5 and 9, FoxA1 can retain its binding capability with DNA, similarly as no ligands are introduced. With both methods, both ligands 1 and 2 are classified as strong inhibitors, ligands 6–8 as weak inhibitors (partial FoxA1 binding can be detected), the rest as non-inhibitor.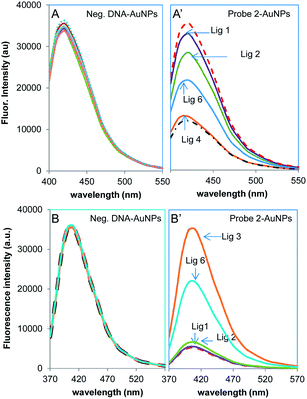 | ||
| Fig. 6 Detection of ligand effect on FoxA1 binding to Probe 2–AuNPs using (A & A′) ACP-430/AuNPs and (B & B′) CCP-410/AuNPs sensors. A negative DNA (mtR3)–AuNPs was used as a control. | ||
Conclusions
Numerous research efforts have been focusing on metal NPs owing to their unique surface Plasmon resonance properties. Both colorimetric and fluorometric methods can help in elucidating biological process and be used as biosensors. In this study, for the first time we coupled dsDNA–AuNP and water soluble conjugated polymers (CPs) for studying protein–dsDNA interaction and ligand inhibition for two important oncogenic factors, i.e. FoxA1 and AP-2γ that are pivotal in regulating the transcriptional activity of estrogen receptor alpha and controlling the expression of estrogen-responsive breast cancer cells. Owing to CP's high quantum yield, charge and AuNPs' super quenching property, dsDNA–AuNP and CP assembly are apposite to explicate the binding affinity of positively charged protein to DNA (and ligand inhibition), either via fluorescence quenching or recovery methods, depending on the CP's charge properties and their spectrum overlay with AuNPs' absorption. In the case when protein's charge property is unknown, it is advisable to utilize CP that is partially quenched in the presence of dsDNA–AuNP entity, because either increase or decrease in fluorescent intensity can be explored. With the versatile hybrid sensors, we have determined the DNA binding properties of FoxA1 and AP-2γ, in terms of sequence selectivity, binding constant, and stoichiometry. These are all important characteristics affecting subsequent transcriptional processes. The identification of inhibitors as drug candidates targeting these two transcription factors could be an alternative in treating breast cancer, in particular those that have become endocrine resistance.Experimental section
Materials. HAuCl4·3H2O (99.99%) and Trisodium citrate dihydrate (99.9%) were obtained from Aldrich Pte Ltd. HisMBP FoxA1 protein and HisMBP AP-2γ were purified in our lab using the protocol as described in ESI.† Both FoxA1 and AP-2γ were in 100 μM stock in 100 mM NaCl, 10 mM Tris–HCl, 2 mM TCEP. For long term storage, the proteins were kept as 10 μL aliquots at −80 °C. Before use, they were quickly thawed in room temperature water bath and returned to 4 °C to maintain the activity.Oligo nucleotides (sense and antisense sequences, Table 1) were purchased from Sigma Life Science. For FoxA1, three DNA probes of different affinity (named Probe 1, Probe 2, and Probe 3) were studied. Probe 2 contains a core FoxA1 site (5′-TGTAAATAAA-3′), while Probe 1 and Probe 3 are modified at the core sequence and the flanking region, respectively. For AP-2γ, to demonstrate its affinity to GC-consensus, one GC rich DNA denoted as wild-type (wtR3) and one GC poor DNA called mutated-type (mtR3) were used for this study. For conjugation of the DNA, these sense sequences are thiol-labeled at the 5′ end for conjugation to AuNPs.
For studying ligand interruption of FoxA1 binding to DNA, 9 small ligands (Table 4) procured from the NCI (National Cancer Institute) compound library were used in this study. All of the ligands were dissolved in DMSO. Before using, they were thawed in a room temperature water bath.
Three water soluble CPs were utilized, two anionic CPs (ACPs), i.e. poly[9,9-bis(40-sulfonatobutyl)fluorene-co-alt-1,4-phenylene] sodium salt (PFP–SO3Na) and poly[5-methoxy-2-(3-sulfopropoxy)-1,4-phenylenevinylene] potassium salt and one cationic CP (CCP), poly[(2,5-bis(2-(N,N-diethylammoniumbromide)ethoxy)-1,4-phenylene)-alt-1,4-phenylene]. According to their emission maximum and charge properties, they are denoted as ACP-430, ACP-560, and CCP-410, respectively. The chemical structures of these CPs are given in Fig. 2A. The ACP-560 and CCP-410 were purchased from Sigma Aldrich, while the anionic-430CP was provided by Prof. Liu Bin (National University of Singapore). This polymer has been previously using for detecting lysozyme using organic dye–CPs FRET.28
FoxA1 and AP-2γ purification
FoxA1 was prepared as HisMBP-tagged FoxA1 recombinant proteins. Full-length FoxA1 cDNA (ESI, Fig. S6†) was cloned into a pHISMBP (Addgene) expression vector via the Gateway cloning system (Invitrogen) as described by the manufacturer. FoxA1 was expressed as a HisMBP fusion protein in BL21 (DE3) cells at 18 °C for 18 h of 0.5 mM IPTG induction in TB (Terrific Broth) media. Cells were collected by centrifugation, resuspended in a lysis buffer containing 50 mM Tris pH 8.0, 300 mM NaCl, 30 mM imidazole and sonicated on ice. Fusion proteins were initially purified from cell lysates using a nickel column equilibrated with the lysis buffer and eluted with the same buffer supplemented with 300 mM imidazole. A second purification step using ion exchange chromatography was performed by passing the sample through a Resource Q anion exchanger (GE Healthcare) and eluted in buffer containing 10 mM Tris–HCl, 1 M NaCl, pH 8.0, with a linear gradient from 0.1–1.0 M NaCl. Eluted fractions were collected, pooled, dialyzed against storage buffer (10 mM Tris–HCl, 100 mM NaCl, 2 mM TCEP, pH 8.0) and concentrated to approximately 100 μM using Vivaspin 20 concentrator before storing at −80 °C until use.Protein purification of HisMBP AP-2γ protein is similar as that for HisMBP–FoxA1 protein till nickel affinity purification step, except that all buffers are at pH 7.0. The HisMBP–AP-2γ protein was subsequently purified using the Hi-Trap Heparin HP column (GE healthcare) with A1 (10 mM HEPES, 100 mM NaCl; pH 7.0) and B1 (10 mM HEPES, 1 M NaCl; pH 7.0) buffers. Eluted fractions containing the fusion protein are pooled together, concentrated in a 30 kDa concentrator and desalted twice in A4 buffer. Protein was then stored at −80 °C in 100 μM aliquots.
Fluorescent anisotropic assay
To determine the binding activity of His–MBP–FoxA1 to various FoxA1 probes containing its cognate DNA element, fluorescence anisotropy assay was utilized. The sequences of the two wild-type and a mutant FAM labeled probes used were as described earlier. The assays were carried out in 384-well microplates (Corning) in which varying concentration of protein were incubated with 2 nM labeled probe in a PBS buffer for 20 min at room temperature (RT). For examining the effects of small molecule inhibitors on FoxA1 DNA binding, 125 nM of protein was pre-incubated with ∼5% DMSO, or 1 μM unlabeled probe, or 1.9 μM inhibitor for 1 h, RT, before the addition of 2 nM FAM labeled probe for another hour incubation. Fluorescence anisotropy was measured using Synergy 2 Multi-Mode Microplate Reader with a 485/20-excitation and a 528/20-emission filter. The equilibrium dissociation constants for each wild-type probe were subsequently calculated by fitting the FoxA1 concentration vs. fluorescence polarization plot using Langmuir isotherm in Origin Pro 8 software. Standard error values (precision of the fitted values) are also obtained from the software after fitting (ESI, Fig. S3†).Synthesis of 13 nm AuNPs and preparation of dsDNA conjugated AuNPs (dsDNA–AuNPs)
AuNPs were synthesized via citrate reduction method, following the procedures available elsewhere.9,10,31 The resulting AuNPs is approximately 13 nm in diameter and in a concentration of 5.33 nM, calculated according to Beer's law, using the extinction coefficient of 2.467 × 108 M−1 cm−1 for 13 nm AuNPs.Thiolated single stranded DNA (ssDNA, i.e. the sense strand) was activated with tris(2-carboxyethyl)phosphine (TCEP, 10 molar excess) and stirred for 10 min. The final solution was centrifuged, with a Sigma-Aldrich microcon centrifugal filter device, YM-3 (NMWCO 3 kDa), to remove TCEP before conjugation to AuNPs. Conjugation of activated thiolated DNA to AuNP was done as described by Zhang et al.32 Activated DNA was mixed with AuNPs at the desired ratio (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and incubated for 5 min. The pH of solution was then lowered to 3 and salt concentration was increased to 30 mM by adding HCl and NaCl, respectively. After 20 min, NaOH was added to return pH to a neutral range. The ssDNA–AuNPs conjugates were then annealed with its complimentary DNA strands at 90 °C for 5 min and let to cool down to room temperature (RT). The amount of bound dsDNA was quantified by thiazole orange staining method as described elsewhere32 and customized for the current study (detail in ESI, Fig. S7†).
1) and incubated for 5 min. The pH of solution was then lowered to 3 and salt concentration was increased to 30 mM by adding HCl and NaCl, respectively. After 20 min, NaOH was added to return pH to a neutral range. The ssDNA–AuNPs conjugates were then annealed with its complimentary DNA strands at 90 °C for 5 min and let to cool down to room temperature (RT). The amount of bound dsDNA was quantified by thiazole orange staining method as described elsewhere32 and customized for the current study (detail in ESI, Fig. S7†).
The hydrodynamic size and zeta potential of AuNPs before and after conjugation with dsDNA, as well as those after protein and CPs binding, were measured with a Dynamic Light Scattering System (BI-200SM, Brookhaven Instruments Corporation). The measurements were performed in water for AuNPs and dsDNA-coated AuNPs, and in 5 mM HEPES buffer for protein and CPs bound NPs.
CPs/DNA–AuNPs hybrid assembly for detecting protein–DNA binding and ligand inhibition
The protein–DNA binding assay was conducted in three main steps. Firstly, 50 nM dsDNA–AuNPs (referring to dsDNA concentration on the surface of AuNPs) was incubated with protein for 20–40 min at RT. To determine the binding constant (Kd) and the stoichiometry, protein titration was conducted with increasing concentration from 0–250 nM. Secondly, CP of 100 nM (i.e. 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio to dsDNA–AuNP) was added to the dsDNA–AuNPs–protein mixture and incubated for an additional 10 min to let the system to reach equilibrium. Finally, the fluorescence spectra of the final solution were measured and compared to a control without any protein. Depending on the charge of the protein and the CPs used, the protein–DNA binding may lead to further fluorescence quenching or fluorescence restore (see Discussion below).
1 molar ratio to dsDNA–AuNP) was added to the dsDNA–AuNPs–protein mixture and incubated for an additional 10 min to let the system to reach equilibrium. Finally, the fluorescence spectra of the final solution were measured and compared to a control without any protein. Depending on the charge of the protein and the CPs used, the protein–DNA binding may lead to further fluorescence quenching or fluorescence restore (see Discussion below).
For screening ligand inhibition, the ligand and protein were incubated (at molar ratio of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 10–20 min at RT prior to adding into DNA-conjugated AuNPs. Subsequently, respective CP was added to the mixture and incubated for 10 min before fluorescence spectra were measured.
1) for 10–20 min at RT prior to adding into DNA-conjugated AuNPs. Subsequently, respective CP was added to the mixture and incubated for 10 min before fluorescence spectra were measured.
Acknowledgements
SX and EC would like to acknowledge the Agency for Science, Technology and Research (A*STAR), Singapore for the financial support (JCO 1131CFG001).References
- S. Su, X. L. Zuo, D. Pan, H. Pei, L. H. Wang, C. H. Fan and W. Huang, Nanoscale, 2013, 5, 2589–2599 RSC
.
- S. Zeng, K. T. Yong, I. Roy, X. Q. Dinh, X. Yu and F. Luan, Plasmonics, 2011, 6, 491–506 CrossRef CAS
.
- K. E. Fong and L. Yung, RSC Adv., 2012, 2, 5154–5163 RSC
.
- K. E. Fong and L. Yung, RSC Adv., 2013, 3, 6076–6084 RSC
.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed
.
- S. P. Song, Y. Qin, Q. Huang, C. H. Fan and H. Y. Chen, Chem. Soc. Rev., 2010, 39, 4234–4243 RSC
.
- J. Wang and X. G. Qu, Nanoscale, 2013, 5, 3589–3600 RSC
.
- Y. N. Tan, X. D. Su, E. Liu and J. S. Thomsen, Anal. Chem., 2010, 82, 2759–2766 CrossRef CAS PubMed
.
- Y. N. Tan, K. H. Lee and X. Su, Anal. Chem., 2011, 83, 4251–4257 CrossRef CAS PubMed
.
- Y. N. Tan, X. D. Su, Y. Zhu and J. Y. Lee, ACS Nano, 2010, 4, 5101–5110 CrossRef CAS PubMed
.
- L. J. Ou, P. Y. Jin, X. Chu, J. H. Jiang and R. Q. Yu, Anal. Chem., 2010, 82, 6015–6024 CrossRef CAS PubMed
.
- J. Fang, L. L. Yu, P. Gao and Y. A. Wei, Anal. Biochem., 2010, 399, 262–267 CrossRef CAS PubMed
.
- C. Zhu, L. B. Liu, Q. Yang, F. Lv and S. Wang, Chem. Rev., 2012, 112, 4687–4735 CrossRef CAS PubMed
.
- X. L. Feng, L. B. Liu, S. Wang and D. B. Zhu, Chem. Soc. Rev., 2010, 39, 2411–2419 RSC
.
- D. F. Moyano, S. Rana, U. H. F. Bunz and V. M. Rotello, Faraday Discuss., 2011, 152, 33–42 RSC
.
- A. Bajaj, O. R. Miranda, I. B. Kim, R. L. Phillips, D. J. Jerry, U. H. Bunz and V. M. Rotello, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10912–10916 CrossRef CAS PubMed
.
- R. L. Phillips, O. R. Miranda, C. You, V. M. Rotello and U. H. Bunz, Angew. Chem., Int. Ed., 2008, 47, 2590–2594 CrossRef CAS PubMed
.
- L. Shang, C. Qin, T. Wang, M. Wang and S. J. J. Dong, Phys. Chem., 2007, 111, 13414–13417 CAS
.
- F. Han, Z. P. Guan, T. S. Tan and Q. H. Xu, ACS Appl. Mater. Interfaces, 2012, 4, 4746–4751 CAS
.
- Z. P. Guan, L. Polavarapu and Q. H. Xu, Langmuir, 2010, 26, 18020–18023 CrossRef CAS PubMed
.
- L. Polavarapu, V. Mamidala, Z. Guan, W. Ji and Q. H. Xu, Appl. Phys. Lett., 2012, 100, 023106 CrossRef PubMed
.
- F. Xia, X. Zuo, R. Yang, Y. Xiao, D. Kang, A. Vallisle, X. J. Gong, D. Yuen, B. B. Hsu and A. J. Heeger, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10837–10841 CrossRef CAS PubMed
.
- J. S. Carroll, X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, W. E. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver and M. Brown, Cell, 2005, 122, 33–43 CrossRef CAS PubMed
.
- S. K. Tan, Z. H. Lin, C. W. Chang, V. Varang, K. R. Chng, Y. F. Pan, E. L. Yong, W. K. Sung and E. Cheung, EMBO J., 2011, 30, 2569–2581 CrossRef CAS PubMed
.
- S. Chatterjee, J. B. Lee, N. V. Valappil, D. Luo and V. M. Menon, Biomed. Opt. Express, 2011, 2, 1727–1733 CrossRef CAS PubMed
.
- H. S. Rye, S. Yue, D. E. Wemmer, M. A. Quesada, R. P. Haugland, R. A. Mathies and A. N. Glazer, Nucleic Acids Res., 1992, 20, 2803–2812 CrossRef CAS PubMed
.
- P. V. Hornbeck, J. M. Kornhauser, S. Tkachev, B. Zhang, E. Skrzypek, B. Murray, V. Latham and M. Sullivan, Nucleic Acids Res., 2012, 40, D261–D270 CrossRef CAS PubMed
.
- J. Wang and B. Liu, Chem. Commun., 2009, 2284–2286 RSC
.
- D. G. Hu, D. Gardner-Stephen, G. Severi, P. A. Gregory, J. Treloar, G. G. Giles, D. R. English, J. L. Hopper, W. D. Tilley and P. I. Mackenzie, Mol. Pharmacol., 2010, 78, 714–722 CrossRef CAS PubMed
.
- T. Williams and R. Tjian, Genes Dev., 1991, 5, 670–682 CrossRef CAS
.
- G. Frens, Nature, 1973, 241, 20–22 CAS
.
- X. Zhang, M. R. Servos and J. Liu, J. Am. Chem. Soc., 2012, 134, 7266–7269 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Protein purification procedure, quantification of dsDNA conjugated to AuNP, fluorescence spectra of ACP-430 in solvent of differing polarity, Fluorescence of CPs in the presence of dsDNA or protein, but not AuNPs, detecting Ap-2γ–DNA interactions using CCP-410/AuNPs and ACP-430/AuNPs hybrid sensors, quantification of Ap-2γ–DNA binding constant (Kd) and stoichiometry (n) through titration. See DOI: 10.1039/c3ra46752j |
| This journal is © The Royal Society of Chemistry 2014 |