Synthesis and discovery of andrographolide derivatives as non-steroidal farnesoid X receptor (FXR) antagonists†
Abstract
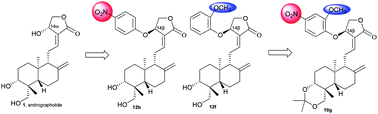
Based upon the discovery of the natural compound andrographolide (1) as a non-steroidal farnesoid X receptor (FXR) antagonist, a series of andrographolide derivatives were designed and synthesized accordingly. Our primary SAR studies demonstrated that 14-phenoxy andrographolide scaffold is an excellent structural pharmacophore for FXR antagonists. Remarkably, 14β-compounds of 12b, 12f and 10g were found to be the most potent FXR antagonists in this work. Structural docking discovered that the phenoxy substitution at the 14-position and the modification at 3,19-positions altered the putative binding positions of small FXR ligands, resulting in their FXR antagonistic activity discrepancy.

 Please wait while we load your content...
Please wait while we load your content...