On-chip detection of multiple serum antibodies against epitopes of celiac disease by an array of amorphous silicon sensors†
Abstract
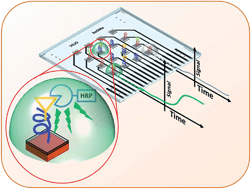
In this paper, we present the preliminary results of an ELISA-on-chip device, intended as a technological demonstrator of a novel analytical system suitable for the diagnosis and follow-up of celiac disease. The idea of the work is to combine an array of amorphous silicon photosensors with a pattern of a poly(2-hydroxyethyl methacrylate) polymer brush film, which acts as anchor for the immobilization of gliadin peptides containing the celiac disease epitopes. Recognition relies on a sandwich immunoassay between antibodies against the peptides and secondary antibodies marked with horseradish peroxidase to obtain a chemiluminescent signal. Detection is based on the measurement of photocurrent induced in the array of amorphous silicon photosensors by the chemiluminescent signal. An ad-hoc procedure has been developed in order to enable the fabrication of the photodiode array and the polymer brush pattern on the two sides of the same glass substrate ensuring the compatibility of the different technological steps. The sensitivity and the selectivity of the chip for multiplex immunoassays were demonstrated using two gliadin peptides (VEA and DEC). In particular, we found that the average amount of the bound HRP revealed by our analytical protocol is 3.5(±0.3) × 10−6 pg μm−2 and 0.85(±0.3) × 10−6 pg μm−2 for specific and non-specific interactions, respectively.

 Please wait while we load your content...
Please wait while we load your content...