Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The study of polyplex formation and stability by time-resolved fluorescence spectroscopy of SYBR Green I-stained DNA
Cosimo
D'Andrea
*abc,
Daniele
Pezzoli
d,
Chiara
Malloggi
e,
Alessia
Candeo
a,
Giulio
Capelli
a,
Andrea
Bassi
ac,
Alessandro
Volonterio
e,
Paola
Taroni
ac and
Gabriele
Candiani
*def
aDepartment of Physics, Politecnico di Milano, Piazza Leonardo da Vinci 32, Milano, 20133, Italy. E-mail: cosimo.dandrea@polimi.it; Tel: +39-02-2399-6113
bCenter for Nano-Science and Technology @ POLIMI, Istituto Italiano di Tecnologia, via G. Pascoli 70/3, 20133 Milano, Italy
cIstituto di Fotonica e Nanotecnologie – Consiglio Nazionale delle Ricerche (IFN-CNR), Piazza Leonardo da Vinci 32, 20133 Milano, Italy
dResearch Unit Milano Politecnico, National Interuniversity Consortium of Materials Science and Technology – INSTM, via Mancinelli 7, 20131 Milano, Italy
eDepartment of Chemistry, Materials, and Chemical Engineering “G. Natta”, Politecnico di Milano, via Mancinelli 7, 20131 Milano, Italy. E-mail: gabriele.candiani@polimi.it; Tel: +39-02-2399-3181
fCentro Interuniversitario di Ricerca in Biotecnologie Proteiche “The Protein Factory”, Politecnico di Milano, CNR–ICRM Milano, and Università degli Studi dell'Insubria, via Mancinelli 7, 20131 Milano, Italy
First published on 18th September 2014
Abstract
Polyplexes are nanoparticles formed by the self-assembly of DNA/RNA and cationic polymers specifically designed to deliver exogenous genetic material to cells by a process called transfection. There is a general consensus that a subtle balance between sufficient extracellular protection and intracellular release of nucleic acids is a key factor for successful gene delivery. Therefore, there is a strong need to develop suitable tools and techniques for enabling the monitoring of the stability of polyplexes in the biological environment they face during transfection. In this work we propose time-resolved fluorescence spectroscopy in combination with SYBR Green I-DNA dye as a reliable tool for the in-depth characterization of the DNA/vector complexation state. As a proof of concept, we provide essential information on the assembly and disassembly of complexes formed between DNA and each of three cationic polymers, namely a novel promising chitosan-graft-branched polyethylenimine copolymer (Chi-g-bPEI), one of its building block 2 kDa bPEI and the gold standard transfectant 25 kDa bPEI. Our results highlight the higher information content provided by the time-resolved studies of SYBR Green I/DNA, as compared to conventional steady state measurements of ethidium bromide/DNA that enabled us to draw relationships among fluorescence lifetime, polyplex structural changes and transfection efficiency.
Introduction
The recent complete mapping of the human genome and sequencing of genes by the Human Genome Project (HGP) have opened new perspectives towards understanding the aetiology and the pathophysiology of human diseases.1 Concomitant advances in materials science and biotechnology have enabled the development of systems for the delivery of therapeutic nucleic acids, and paved the way for gene therapy strategies to treat a variety of diseases.Although the ease of synthesis, relatively low cost and high safety make cationic polymers attractive and valuable tools for the delivery of nucleic acids to cells,2 their widespread use in clinical trials has mainly been hampered by transfection efficiency concerns. These gene delivery vectors are inherently cationic at physiological pH, spontaneously self-organizing with polyanionic nucleic acids in nano- and microparticles named polyplexes.3,4 In this scenario, there is a growing consensus that a better understanding of the processes leading to DNA complexation/release and finally to effective polyfection represents a big step forward in the rational design of effective polymeric gene vectors.
Polyethylenimine (PEI) is considered as the gold standard gene carrier among polymeric non-viral vectors. It exists in either linear (lPEI) or branched (bPEI) form and, although conflicting results are reported in the literature, several authors have pointed out the superior transfection efficiency of bPEI.5
Both the transfection efficiency and cytotoxicity of PEIs are strictly related to their molecular weight (Mw): high Mw (HMW) PEIs not only lead to enhanced transfection but also induce undesirable cytotoxicity.6,7 Recently, aiming to reduce the toxic side effects of HMW PEIs while preserving their superior transfection ability, low Mw (LMW) bPEI has been grafted onto medium Mw (MMW) chitosan, to obtain chitosan-graft-bPEI (Chi-g-bPEI) copolymers featuring lower cytotoxicity and equal or higher transfection efficiency than that of the gold standard transfectant 25 kDa bPEI.8–10 However, the reason(s) why these polymers display such different behaviours has been overlooked thus far.
Due to the excellent sensitivity of fluorescence compared to other optical techniques and to the large variety of fluorescent intercalating dyes available in the market, spectrofluorimetric steady-state analyses of DNA-dyes such as ethidium bromide (EtBr) are usually performed in fluorophore-exclusion assays to investigate the DNA-condensation capability of gene delivery vectors.11–13 However, this technique does not allow discrimination between the expulsion of the dye from the DNA double helix and possible changes in its fluorescence quantum yield due to e.g. DNA rearrangement internal to polyplexes.
Recently, the use of time-resolved fluorescence spectroscopy techniques in combination with the DNA-intercalant EtBr was proposed as a feasible analytical tool to study the state of nucleic acids within polyplexes.14–17 Time-resolved techniques indeed allow disentangling the fluorescence amplitude and lifetime, thus providing additional information with respect to popular steady-state analyses. In fact, the lifetime of fluorescent DNA-dyes, and of every fluorophore in general, is strongly dependent on the microenvironment and, hence, on the binding strength between the polymer and the DNA in the case of polyplexes. Transfection effectiveness of polyplexes arises from the suitable balance between the extracellular stability and the ability to release their cargo inside the cells. Therefore, a direct measurement of the DNA complexation state in the different environments resembling those encountered by polyplexes in living systems is fundamental to characterize their behaviour.
To the best of our knowledge the present work is the first to present the use of SYBR Green I in combination with time-resolved fluorescence spectroscopy to investigate the DNA condensation state in polyplexes. This issue is also of particular importance to the large segment of the research focused on gene regulation and theranostic applications. Furthermore, due to its superior safety profile and fluorescence properties, SYBR Green I has superseded EtBr in most laboratories soon after its launch on the market. We have thus evaluated the fluorescence decay of SYBR Green I/DNA in polyplexes during the assembly and disassembly steps in biological media, comparing the behaviour of three different cationic polymers: 2 kDa bPEI, 25 kDa bPEI and a Chi-g-bPEI copolymer. Overall, the main aim of our study was to find out the possible relationship among the optical behaviour, physicochemical properties and transfection profiles of cationic polymers.
The strong fluorescence signal of SYBR Green I/DNA and its high information content together with the increasingly widespread use of time-resolved fluorescence spectroscopy and the typical advantages of optical techniques, such as high sensitivity, speed to follow dynamic processes, easy-to-use instrumentation and cost-effectiveness, make us confident in the routine use of the technique described herein in (bio)chemical laboratories for the development of non-viral gene delivery vectors.
Results and discussion
Recently, time-resolved fluorescence in combination with EtBr was used to compare the DNA-complexation behaviour of various polymeric gene delivery vectors.14,16 Unfortunately, possibly owing to the too low signal of EtBr, fluorescence measurements had to be performed at a very high DNA concentration (300 μM, corresponding to ≈0.1 μg μL−1) far above those generally used in vitro (on the order of 1–10 ng μL−1 of DNA or even lower),10,18 and obviously in vivo, in gene delivery assays, inasmuch as certain suspensions were reported to be cloudy.In light of this drawback, we introduced herein the use of SYBR Green I as the DNA-dye for time-resolved fluorescence spectroscopy experiments on polyplexes. Both the fluorescence quantum yield and fluorescence enhancement in going from the SYBR Green I free dye to the double-stranded DNA (dsDNA) intercalated dye are much higher than for EtBr,19 thus allowing the study of the DNA behaviour in polyplexes at realistic concentrations. Specifically, we compared the DNA-complexation behaviour of a Chi-g-bPEI copolymer with a degree of grafting of 6.5% onto those of 2 kDa and 25 kDa bPEI, to find out the possible relationship with the physicochemical and biological properties of the corresponding polyplexes. It is worth recalling that 2 kDa bPEI is the transfection-active building block of the Chi-g-bPEI copolymer, while 25 kDa bPEI is a gold standard transfectant.
Time-resolved fluorescence of polyplexes in the presence of SYBR Green I
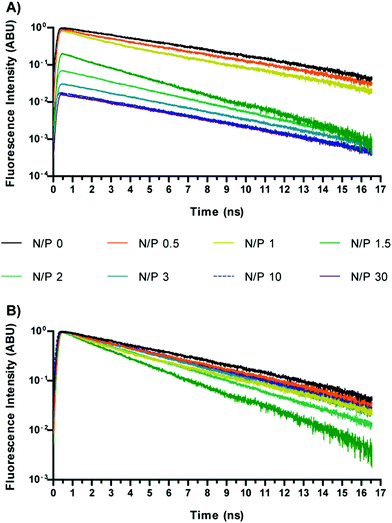
Preliminary measurements were carried out in order to estimate the fluorescence lifetime of SYBR Green I in the absence and in the presence of DNA. Free SYBR Green I displayed an extremely low signal and a short temporal decay, below the temporal resolution of the system (≈200 ps). The low quantum yield of free SYBR Green I is in good agreement with a previous report by Cosa and co-workers.19 Instead, no significant increase of the fluorescence signal was detected upon mixing SYBR Green I with each polymer investigated (data not shown). On the other hand a tremendous increase in fluorescence intensity was noted when SYBR Green I was kept in the presence of plain DNA, the decay showing a mono-exponential behaviour with a lifetime of ca. 5.5 ns.Fig. 1 shows the time-resolved fluorescence curves for Chi-g-bPEI polyplexes prepared at different nitrogen to phosphate ratios (N/P) and diluted in 10 mM Hepes pH 7. The wavelength selected in these and all the forthcoming results was 530 ± 10 nm, which is close to the maximum emission of SYBR Green I. This choice was motivated by the experimental evidence that unlike EtBr that showed temporal and spectral variations following DNA condensation by polymers,14 no change occurred in the emission spectrum of SYBR Green I upon variation of the N/P (not shown). This aspect supports the approach of focusing on the lifetime of SYBR Green I to get insight into the polyplex conformation and stability. As expected, we also observed a strong reduction in the SYBR Green I fluorescence amplitude upon increasing N/P (Fig. 1), owing to the progressive removal of the dye from nucleic acids. Most importantly, we observed a change in the fluorescence lifetime, indicating the variation of the quantum yield of the dye still bound to the DNA. A similar behaviour was also observed for 2 kDa and 25 kDa bPEI, as shown in Fig. 2.
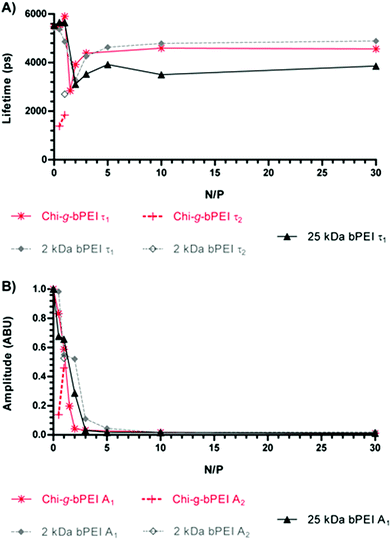
In order to quantify the fluorescence amplitudes and lifetimes, the time profiles corresponding to different N/Ps were generally best-fitted to a bi-exponential function, except for some cases in which the fitting procedure converged towards a single exponential decay. Fluorescence amplitudes and lifetime values as a function of the N/P for the three polymers are reported in Fig. 2.
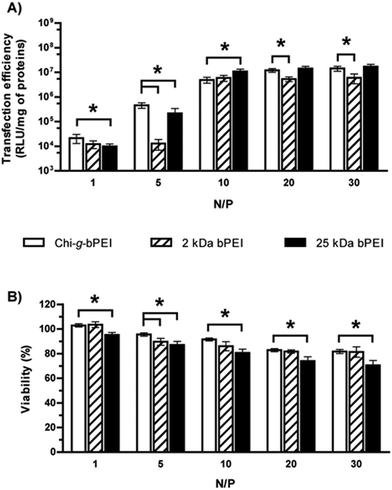
The temporal behaviour of the emitted fluorescence was anomalous for N/P ≤ 2. Indeed, the curves showed a bi-exponential profile and/or a strong reduction of longer lifetimes. Upon increasing the N/P, the temporal profiles always went back to a mono-exponential function and the fluorescence lifetimes began to increase, though they never reached the maximum value recorded in the presence of naked DNA (N/P 0). Due to the strong dependence of the fluorescence lifetime on the microenvironment,20–22 these changes can be related to different conformational states of the DNA as a function of the N/P. Furthermore, even at high N/P, SYBR Green I was not completely expelled from the double helix and a reduced but significant amount of fluorescent dye was still adsorbed or laid in close proximity to the nucleic acids, representing a valuable marker to unravel the conformation of polyplexes and to study their stability in biological environments. In fact, although to a lesser extent, the lifetime changes observed for those N/Ps identified as the most effective in transfection experiments (N/P ≥ 10, Fig. 4) pointed out dynamic variations in the polyplex structure.23 Of note, transfections were carried out with the most widely used standard, i.e. the luciferase-encoding plasmid and transfection efficiency expressed as relative luciferase units per milligram of cell protein lysate (RLU mg−1 of proteins).10,24
In an attempt to shed light on the possible relationship between the fluorescence lifetime and the polyplex structure, the average hydrodynamic diameter (DH) (Fig. 3A) and ζ-potential (ζP) (Fig. 3B) of the polyplexes at varying N/Ps were measured by dynamic light scattering (DLS) and laser Doppler micro-electrophoresis, respectively. At low N/Ps (N/P < 1.5) the surface charge of DNA/Chi-g-bPEI complexes was negative probably because of the exposure of the phosphates on the outside of polyplexes (i.e. ineffective DNA shielding). At N/P 1.5 Chi-g-bPEI-based polyplexes displayed a ζP close to neutrality and their biggest size (DH ≈ 1200 nm); this can be ascribed to aggregation of complexes in the absence of net electrostatic repulsive forces.9,10 An increase in fluorescence quenching would be expected in such a condition and this is in good agreement with time-resolved fluorescence measurements. In particular, we observed a bi-exponential behaviour showing a fluorescence lifetime reduction of the longer component and the appearance of a second short component, which can reasonably be ascribed to the aggregation of the complexes. Moreover, as the N/P was further augmented, the electrostatic repulsive forces prevented the aggregation of polyplexes and caused the total embedding of the DNA in nanometric polyplexes. Afterwards, the size of the particles remained fairly constant while the fluorescence lifetime of SYBR Green I was still changing so that we could appreciate different DNA conformational states.
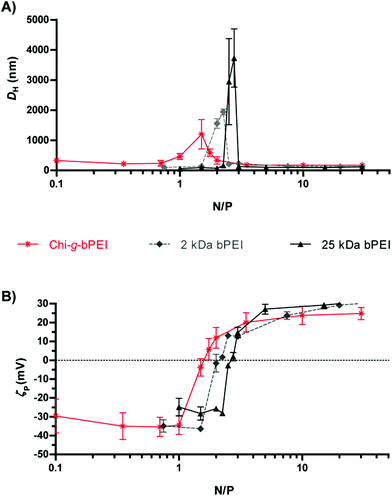 | ||
| Fig. 3 (A) Size (average hydrodynamic diameter, DH) and (B) surface charge (ζ-potential, ζP) of Chi-g-bPEI, 2 kDa and 25 kDa bPEI at varying nitrogen to phosphate ratios (N/Ps). | ||
Although dimensionally different, polyplexes constituted of 2 kDa and 25 kDa bPEI showed qualitatively similar profiles with an anomalous behaviour of the fluorescence lifetime in correspondence with the null surface potential (0 mV) (Fig. 3B) and with their respective maxima in size (Fig. 3A) (N/P ≈ 2). At N/P > 2, the fluorescence lifetimes of 2 kDa bPEI and Chi-g-bPEI complexes were instead higher compared to that of 25 kDa bPEI (Fig. 2A). Hence, the latter is possibly characterized by a higher DNA binding strength. This could account for the reduction of the fluorescence quantum yield (and consequently of the lifetime) and for the greater displacement of the SYBR Green I from the DNA. However, its removal was never complete, as shown by the higher fluorescence intensity and longer lifetime compared to that of the free SYBR Green I dye.
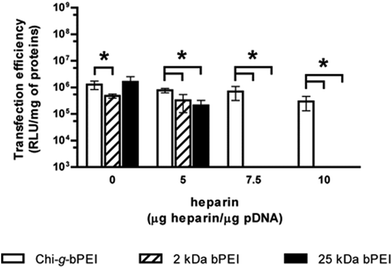
Fig. 4 shows the transfection efficiency for all polyplexes in complete DMEM (defined in the Experimental section). Although all are similar in size and surface charge (Fig. 3), the polymer/DNA complexes showed quite different transfection behaviours, as evidenced by the measurement of luciferase activity. In light of these findings we can speculate that the dimensions and the ζP of polyplexes per se are poorly predictive of the transfection outcome.25 In contrast, each kind of polyplex displayed a peculiar lifetime value that highlighted polymer-specific DNA arrangements.
Concerning the cytotoxicity of gene delivery vectors, similar levels were observed for Chi-g-bPEI- and 2 kDa bPEI-based polyplexes for all the N/Ps tested, apart from N/P 5 (p < 0.05). Irrespective of the N/P, the cytotoxicity of plasmid DNA (pDNA)/25 kDa bPEI complexes was instead the highest.
Interestingly, the fluorescence lifetime value of SYBR Green I/DNA in the presence of the copolymer at N/P > 3 was between those of LMW and HMW bPEI-complexes and closer to that of 2 kDa bPEI. Hence, the lifetime of Chi-g-bPEI seemed to be mainly influenced by the presence of 2 kDa bPEI blocks, although a lifetime change due to its conformation and/or to its more intricate structure was also evident. This observation was further evidenced at high N/P by the larger polyplex dimension (i.e. weaker condensation) in the case of the copolymer in comparison with bPEIs and it would account for its noteworthy performance in transfection.9
Polyplex assembly
In order to gain better insight into the process of polyplex formation, a dynamical time-resolved fluorescence measurement was carried out. The time course of the polyplex assembly lasted for ≈75 min. After monitoring SYBR Green I/DNA (N/P 0) for 1 min, each polymer was injected into the cuvette to give polyplexes at N/P 30; the final DNA concentration was kept constant in all the experiments (0.02 μg μL−1). N/P 30 was chosen owing to the high transfection efficiency of every polymer under these conditions (Fig. 4A). The sampling was carried out every 12 s for the first 12 min, while in the remaining time-lapse the sampling interval was extended to 120 s. Time-resolved profiles always showed a mono-exponential temporal behaviour, as shown in Fig. 5.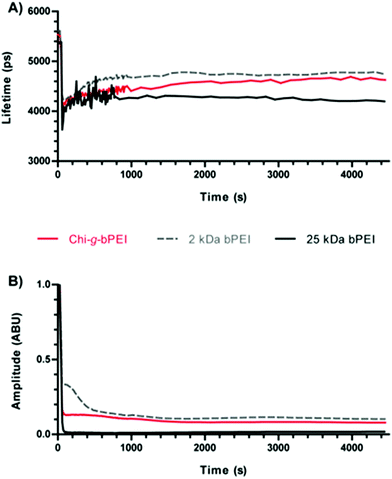 | ||
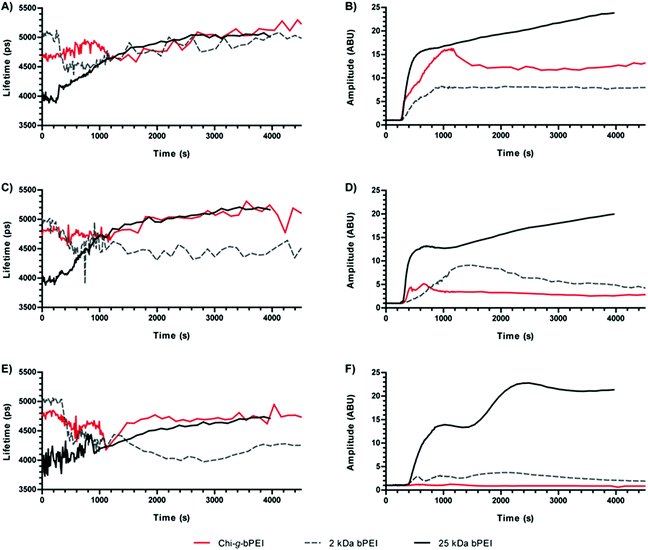
| Fig. 5 (A) Fluorescence lifetime and (B) amplitude of SYBR Green I/DNA as a function of time for Chi-g-bPEI, 2 kDa and 25 kDa bPEI polyplexes at a nitrogen to phosphate ratio (N/P) of 30. | ||
Once every polymer was added to the SYBR Green I/DNA, a dramatic reduction of the fluorescence amplitude was observed: 25 kDa bPEI showed the most drastic decrease to very low residual fluorescence levels (≈2%), while 2 kDa bPEI and Chi-g-bPEI exhibited significant ≈10% and 7% residual fluorescence signals, respectively, even after long-lasting observation (Fig. 5B). Moreover, we observed variations in the fluorescence lifetime over 1500 s for both bPEIs and 2000 s for Chi-g-bPEI, followed by plateauing (Fig. 5A). Since the lifetime variations can be explained with conformational changes of polyplexes, these results pointed out that bPEIs and Chi-g-bPEI complexes take at least 25 and 35 min, respectively, to mature to the final conformation.
For every kind of polyplex studied the addition of the polymer to the SYBR Green I/DNA led to an early but significant reduction of the fluorescence lifetime that decreased from ≈5.5 ns in the absence of transfectant (N/P 0) to 4.1 ns for Chi-g-bPEI, 4.2 ns for 2 kDa bPEI and 3.7 ns for 25 kDa bPEI. Interestingly, these intermediate states showed lifetime values similar to those observed at N/P 1.5–2. This would be explained by the transient formation of unstable aggregates during the early phases of polyplex maturation characterized by a neutral net electrostatic charge. Afterwards, the lifetime increased until reaching a plateau, in agreement with our findings reported in the previous section. The three polyplexes showed different time constant kinetics while joining the plateau phase: Chi-g-bPEI did display the slowest time constant, while 25 kDa bPEI showed the fastest one. It is worth noting that although the copolymer and its building block 2 kDa bPEI exhibited similar lifetimes at the plateau, they were characterized by different time constants while approaching it (Fig. 5A). This could be ascribed to the more complex chemical structure of the former, thus allowing many diverse intermediate states. Moreover, we observed that both the fluorescence amplitude and the lifetime of 25 kDa bPEI-based complexes were characterized by the highest decrease and the fastest kinetics compared to the other polyplexes. These aspects confirmed our previous findings on the highest binding strength of 25 kDa for the DNA.
Polyplex disassembly
In the previous section we showed the polymer/DNA complexation dynamics. The reverse process, i.e. the polyplex disassembly in the presence of anionic competitors, can profitably be studied to mimic the DNA release and it can thus provide some reliable information on the polyplex stability/instability in the biological environment.20 In order to do so, these experiments were carried out by adding heparin to polyplexes at equilibrium, diluted in 10 mM Hepes pH 7 to reflect the actual working concentration. Polyanionic heparin was selected as a suitable model molecule for glycoconjugates such as proteoglycans (PG) and glycosaminoglycans (GAG) that are abundantly present in serum, on the exofacial side of the plasmalemma and dispersed throughout the extracellular space (ECS) and that all compete with the nucleic acids for cationic polymers, possibly leading to polyplex disruption and eventually triggering their disassembly. By monitoring the disintegration process, these measurements aimed at monitoring the polyplex stability under conditions resembling the cellular and tissular environments. In order to modulate the anionic strength of the medium, three different heparin concentrations were used, as reported in the Experimental section.Similar to the previous section, the decomplexation process was dynamically monitored over a ≈75 min period upon the addition of heparin to each polyplex suspension (at 120 s). In the initial 16 min the sampling interval was 12 s, while it was lengthened to every 120 s thereafter. In concomitance with changes in the sampling rate, the laser intensity was reduced to compensate for the increased signal and data were rescaled accordingly. Fig. 6 shows the temporal behaviour of fluorescence amplitudes and lifetimes at the three different heparin concentrations. Polyplexes were characterized by an increase in the fluorescence amplitude due to the competitive effect of heparin which caused their disassembly and the release of the DNA that became thus accessible to the SYBR Green I in the solution. In order to easily compare these variations, we have normalized the amplitudes before the addition of heparin. 25 kDa bPEI-based complexes exhibited lower resistance to heparin than 2 kDa bPEI and Chi-g-bPEI polyplexes, as evidenced by the huge increase in fluorescence amplitude even after the addition of the lowest amount of heparin (5 μg of heparin μg−1 of DNA) as previously reported.11 Importantly, under these conditions, heparin failed to cause any significant release of DNA from Chi-g-bPEI-polyplexes. This highlighted their greatest stability in an extracellular-mimicking environment (Fig. 6F).
The addition of heparin to polyplexes at the equilibrium induced changes that implied variations in their structures. Upon heparin addition, the fluorescence decays of 2 kDa bPEI- and the copolymer-based polyplexes showed the appearance of a bi-exponential behaviour, in particular at high heparin/DNA ratios, while 25 kDa bPEI preserved a mono-exponential profile during the time-lapse of the measurement. Interestingly, these behaviours are analogous to those observed for polyplexes prepared at low N/Ps (Fig. 2).
Comparing the fluorescence lifetimes at varying heparin concentrations, it is interesting to observe that at the highest heparin concentration (10 μg of heparin μg−1 of pDNA) all the three polyplexes converged to the same lifetime value of ≈5.1 ns, close to that of N/P 0 (≈5.5 ns) (Fig. 6A), confirming the release of DNA shown by the increase in amplitude (Fig. 6B).
Instead, at lower heparin concentrations (7.5 and 5 μg of heparin μg−1 of DNA) the three polyplexes showed very different behaviours, while in the case of 25 kDa bPEI the fluorescence lifetime always increased upon heparin injection, for 2 kDa bPEI-based polyplexes it abruptly decreased toward values similar to those observed in the initial phases of polyplex formation, thus indicating only partial disassembly. It is noteworthy that Chi-g-bPEI-polyplexes were the most stable: the fluorescence amplitude underwent the smallest variation and also the lifetime showed minor changes that highlighted the stable conformation of the polyplexes formed.
In order to find out the possible relationship between these optical parameters and the transfection behaviour, HeLa cells underwent transfection with 2 kDa, 25 kDa bPEI, and Chi-g-bPEI at N/P 30 after preincubation for 30 min in 10 mM Hepes pH 7 containing different concentrations of heparin. As shown in Fig. 7, preincubation of 2 kDa and 25 kDa bPEI-polyplexes with the lowest polyanion concentration (5 μg of heparin μg−1 of DNA) drastically reduced the transfection efficiency while at higher concentrations the sulphated glycosaminoglycan completely blunted transgene expression. In sharp contrast, the transfection efficiency of Chi-g-bPEI-based polyplexes was only slightly affected by the preincubation with the same amounts of heparin as before and still remained elevated at the highest concentration, confirming the superior stability of the copolymer-based polyplexes.
Conclusions
In conclusion, in this work we demonstrated that the fluorescence lifetime of the SYBR Green I intercalated within DNA provides valuable information on the formation/disassembly processes of polyplexes. In this regard, the high fluorescence quantum yield and fluorescence enhancement in going from SYBR Green I free dye to dye-DNA enabled easy monitoring of the condensation state of DNA in polyplexes and a strict evaluation of their stability at working concentrations.By an in-depth analysis of the assembly and the disassembly steps of 2 kDa bPEI-, 25 kDa bPEI-, and Chi-g-bPEI-based polyplexes we identified relationships among the optical behaviour, their physicochemical properties and their transfection efficiencies and we shed light on the excellent transfection properties of Chi-g-bPEI-based polyplexes.9,10 Of note, fluorescence lifetime measurements of SYBR Green I/DNA provided some useful information that goes beyond the simple evaluation of the DNA-complexation ability of a given polymer and the characterization of the polyplex size and surface charge, giving insight into the evolution and the stability of polyplexes in biological media. This information that cannot be obtained by steady state measurements carried out using conventional spectrofluorimeters is extremely useful because this and other studies have demonstrated a direct relationship between the polyplex stability and the transfection performance of non-viral gene vectors both in vitro and in vivo.26–28
Future studies will be carried out on other polyplexes and also on lipoplexes to investigate further the potentiality of the use of time-resolved fluorescence spectroscopy in combination with SYBR Green I as a powerful tool for the in-depth characterization of non-viral transfectants. Furthermore, given the increasing use of SYBR Green I in (bio)chemical laboratories and the rapid development and decreasing cost of commercial fluorescence lifetime benchtop spectrofluorimeters, time-resolved fluorescence spectroscopy coupled to SYBR Green I-stained DNA has the potential to become a widespread technique for the development and characterization of non-viral gene delivery vectors and in general for studies involving changes in the condensation state of DNA.
Experimental
Materials
Luciferase Assay System and the pDNA encoding for the modified firefly luciferase pGL3-Control Vector (5.2 kb) were purchased from Promega (Italy). BCA Protein Assay Kit was from Pierce Chemical (IL, USA). HeLa cells (human cervix carcinoma, CCL-2.2) were purchased from the American Type Culture Collection (ATCC, VA, USA). MMW chitosan (Mw 1.9–3.1 × 105; deacetylation degree: 75–85%; Brookfield viscosity: 200–800 cP, 1% in 1% CH3COOH), 2 kDa bPEI (Mn ≈ 1.8 × 103, Mw ≈ 2.0 × 103), 25 kDa bPEI (Mn ≈ 1.0 × 104, Mw ≈ 2.5 × 104) and all other chemicals were from Sigma-Aldrich (Italy) if not differently specified. The use of a cationic polymer based on PEI for transfection is covered by US Patent 6,013,240, European Patent 0,770,140, and foreign equivalents, for which Polyplus-transfection™ is the worldwide exclusive licensee. SYBR Green I (104× in DMSO) is an unsymmetrical cyanine dye which is an ultrasensitive stain of dsDNA (λex = 490 nm, λem = 530 nm).Methods
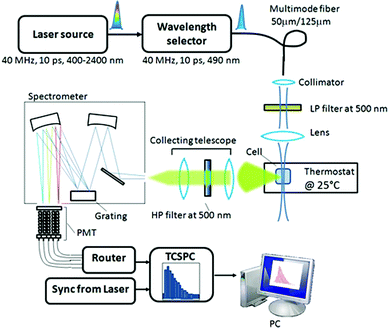 | ||
| Fig. 8 Experimental set-up of fluorescence lifetime measurements. HP: high-pass; LP: low-pass; PMT: photomultiplier tube; TCSPC: Time-Correlated Single Photon Counting. | ||
The fluorescence signal was collected orthogonally to the excitation by a lens system and focused on the entrance slit of an imaging spectrometer (SP-2150i Princeton Instruments, United States). In order to separate the fluorescence signal from the scattered excitation light a high-pass filter with a cut-off at 500 nm was inserted in the optical path between the sample and the spectrometer. The latter was based on a grating with 1200 lines mm−1 and a wavelength blaze at 600 nm. The spectrally dispersed fluorescence light was finally imaged onto a 32-channel photomultiplier tube (H7260 Hamamatsu, Japan). Due to the size of the single photocathode channel (about 1 mm) and the dispersion properties of the spectrometer, a resolution of ca. 4.5 nm per channel was obtained. The detection system relied on the TCSPC technique performed by a data acquisition board (SPC-130 Becker and Hickl, Germany) and a home-made router. The Instrumental Response Function (IRF) was about 200 ps wide (FWHM), as measured by substituting the fluorescent sample with water and detecting the scattered laser light. For fluorescence measurements, pGL3 was mixed with SYBR Green I in 10 mM Hepes buffer pH 7.0 (final concentrations: 0.05 μg μL−1 of pGL3; 20× SYBR Green I) prior to the addition to the polymer solution. Then polyplexes were prepared as described above and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 in 10 mM Hepes buffer pH 7.0.
9 in 10 mM Hepes buffer pH 7.0.
To live-monitor the formation of polyplexes the SYBR Green I/pGL3 suspension was placed in the measurement cuvette and fluorescence signal acquisition was started. After 1 min the polymer solution was added without interrupting the acquisition. The signal was recorded every 12 s in the first 12 min while the time-lapse was extended to 120 s thereafter. The final DNA concentration in the polyplex solution was 0.02 μg μL−1.
To investigate the polymer/DNA disassembly, polyplexes were prepared in the presence of SYBR Green I as described above, hence diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 in 10 mM Hepes buffer pH 7.0, and the fluorescence was recorded for 120 s. Afterwards, without interrupting the acquisition, a concentrated solution of heparin in 10 mM Hepes was added dropwise. The signal was recorded every 12 s in the first 16 min and then extended to every 120 s for a total duration of ≈75 min. The final concentrations of heparin were 0.020 μg μL−1, 0.015 μg μL−1 and 0.010 μg μL−1, corresponding to 10, 7.5 and 5 μg of heparin μg−1 of pDNA, respectively.
9 in 10 mM Hepes buffer pH 7.0, and the fluorescence was recorded for 120 s. Afterwards, without interrupting the acquisition, a concentrated solution of heparin in 10 mM Hepes was added dropwise. The signal was recorded every 12 s in the first 16 min and then extended to every 120 s for a total duration of ≈75 min. The final concentrations of heparin were 0.020 μg μL−1, 0.015 μg μL−1 and 0.010 μg μL−1, corresponding to 10, 7.5 and 5 μg of heparin μg−1 of pDNA, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in 10 mM Hepes buffer pH 7.0 and equilibrated at 25 °C prior to measurements.
10 in 10 mM Hepes buffer pH 7.0 and equilibrated at 25 °C prior to measurements.
For transfection experiments in the presence of heparin, HeLa cells were transfected with 2 kDa bPEI, 25 kDa bPEI and Chi-g-bPEI at N/P 30 after preincubation for 30 min (i) in plain 10 mM Hepes pH 7 or (ii) in 10 mM Hepes pH 7 containing different concentrations of heparin (0.02 μg μL−1, 0.015 μg μL−1 and 0.01 μg μL−1, corresponding to 10, 7.5 and 5 μg of heparin μg−1 of pDNA, respectively). 24 h after transfection, the Alamar Blue assay was performed while the luciferase activity was tested as described above.
Acknowledgements
Research was partially funded by the Italian Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) Futuro in Ricerca (FIRB), grant number: RBFR08XH0H (to CDA and GC) and by Politecnico di Milano, Fondo di Ateneo per la Ricerca di Base (FARB) (to AV and GC).Notes and references
- S. G. Gregory, K. F. Barlow, K. E. McLay, R. Kaul, D. Swarbreck, A. Dunham, C. E. Scott, K. L. Howe, K. Woodfine, C. C. Spencer, M. C. Jones, C. Gillson, S. Searle, Y. Zhou, F. Kokocinski, L. McDonald, R. Evans, K. Phillips, A. Atkinson, R. Cooper, C. Jones, R. E. Hall, T. D. Andrews, C. Lloyd, R. Ainscough, J. P. Almeida, K. D. Ambrose, F. Anderson, R. W. Andrew, R. I. Ashwell, K. Aubin, A. K. Babbage, C. L. Bagguley, J. Bailey, H. Beasley, G. Bethel, C. P. Bird, S. Bray-Allen, J. Y. Brown, A. J. Brown, D. Buckley, J. Burton, J. Bye, C. Carder, J. C. Chapman, S. Y. Clark, G. Clarke, C. Clee, V. Cobley, R. E. Collier, N. Corby, G. J. Coville, J. Davies, R. Deadman, M. Dunn, M. Earthrowl, A. G. Ellington, H. Errington, A. Frankish, J. Frankland, L. French, P. Garner, J. Garnett, L. Gay, M. R. Ghori, R. Gibson, L. M. Gilby, W. Gillett, R. J. Glithero, D. V. Grafham, C. Griffiths, S. Griffiths-Jones, R. Grocock, S. Hammond, E. S. Harrison, E. Hart, E. Haugen, P. D. Heath, S. Holmes, K. Holt, P. J. Howden, A. R. Hunt, S. E. Hunt, G. Hunter, J. Isherwood, R. James, C. Johnson, D. Johnson, A. Joy, M. Kay, J. K. Kershaw, M. Kibukawa, A. M. Kimberley, A. King, A. J. Knights, H. Lad, G. Laird, S. Lawlor, D. A. Leongamornlert, D. M. Lloyd, J. Loveland, J. Lovell, M. J. Lush, R. Lyne, S. Martin, M. Mashreghi-Mohammadi, L. Matthews, N. S. Matthews, S. McLaren, S. Milne, S. Mistry, M. J. Moore, T. Nickerson, C. N. O'Dell, K. Oliver, A. Palmeiri, S. A. Palmer, A. Parker, D. Patel, A. V. Pearce, A. I. Peck, S. Pelan, K. Phelps, B. J. Phillimore, R. Plumb, J. Rajan, C. Raymond, G. Rouse, C. Saenphimmachak, H. K. Sehra, E. Sheridan, R. Shownkeen, S. Sims, C. D. Skuce, M. Smith, C. Steward, S. Subramanian, N. Sycamore, A. Tracey, A. Tromans, Z. Van Helmond, M. Wall, J. M. Wallis, S. White, S. L. Whitehead, J. E. Wilkinson, D. L. Willey, H. Williams, L. Wilming, P. W. Wray, Z. Wu, A. Coulson, M. Vaudin, J. E. Sulston, R. Durbin, T. Hubbard, R. Wooster, I. Dunham, N. P. Carter, G. McVean, M. T. Ross, J. Harrow, M. V. Olson, S. Beck, J. Rogers, D. R. Bentley, R. Banerjee, S. P. Bryant, D. C. Burford, W. D. Burrill, S. M. Clegg, P. Dhami, O. Dovey, L. M. Faulkner, S. M. Gribble, C. F. Langford, R. D. Pandian, K. M. Porter and E. Prigmore, The DNA sequence and biological annotation of human chromosome 1, Nature, 2006, 441, 315–321 CrossRef CAS PubMed.
- G. Y. Wu and C. H. Wu, Receptor-mediated in vitro gene transformation by a soluble DNA carrier system, J. Biol. Chem., 1987, 262, 4429–4432 CAS.
- G. Candiani, D. Pezzoli, L. Ciani, R. Chiesa and S. Ristori, Bioreducible liposomes for gene delivery: from the formulation to the mechanism of action, PLoS One, 2010, 5, e13430 Search PubMed.
- D. Pezzoli and G. Candiani, Non-viral gene delivery strategies for gene therapy: a “menage a trois” among nucleic acids, materials, and the biological environment Stimuli-responsive gene delivery vectors, J. Nanopart. Res., 2013, 15, 1523 CrossRef.
- J. Intra and A. K. Salem, Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo, J. Controlled Release, 2008, 130, 129–138 CrossRef CAS PubMed.
- W. T. Godbey, K. K. Wu and A. G. Mikos, Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle, J. Biomed. Mater. Res., Part A, 1999, 45, 268–275 CrossRef CAS.
- D. Fischer, Y. Li, B. Ahlemeyer, J. Krieglstein and T. Kissel, In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis, Biomaterials, 2003, 24, 1121–1131 CrossRef CAS PubMed.
- K. Wong, G. Sun, X. Zhang, H. Dai, Y. Liu, C. He and K. W. Leong, PEI-g-chitosan, a novel gene delivery system with transfection efficiency comparable to polyethylenimine in vitro and after liver administration in vivo, Bioconjugate Chem., 2006, 17, 152–158 CrossRef CAS PubMed.
- H. L. Jiang, Y. K. Kim, R. Arote, J. W. Nah, M. H. Cho, Y. J. Choi, T. Akaike and C. S. Cho, Chitosan-graft-polyethylenimine as a gene carrier, J. Controlled Release, 2007, 117, 273–280 CrossRef CAS PubMed.
- D. Pezzoli, F. Olimpieri, C. Malloggi, S. Bertini, A. Volonterio and G. Candiani, Chitosan-Graft-Branched Polyethylenimine Copolymers: Influence of Degree of Grafting on Transfection Behavior, PLoS One, 2012, 7, e34711 CAS.
- M. Ruponen, S. Yla-Herttuala and A. Urtti, Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies, Biochim. Biophys. Acta, Biomembr., 1999, 1415, 331–341 CrossRef CAS.
- D. Pezzoli, A. Kajaste-Rudnitski, R. Chiesa and G. Candiani, Lipid-Based Nanoparticles as Nonviral Gene Delivery Vectors, Methods Mol. Biol., 2013, 1025, 269–279 CAS.
- A. Ghilardi, D. Pezzoli, M. C. Bellucci, C. Malloggi, A. Negri, A. Sganappa, G. Tedeschi, G. Candiani and A. Volonterio, Synthesis of Multifunctional PAMAM-Aminoglycoside Conjugates with Enhanced Transfection Efficiency, Bioconjugate Chem., 2013, 24, 1928–1936 CrossRef CAS PubMed.
- E. Vuorimaa, T. M. Ketola, J. J. Green, M. Hanzlikova, H. Lemmetyinen, R. Langer, D. G. Anderson, A. Urtti and M. Yliperttula, Poly(beta-amino ester)-DNA complexes: Time-resolved fluorescence and cellular transfection studies, J. Controlled Release, 2011, 154, 171–176 CrossRef CAS PubMed.
- T. M. Ketola, M. Hanzlikova, A. Urtti, H. Lemmetyinen, M. Yliperttula and E. Vuorimaa, Role of polyplex intermediate species on gene transfer efficiency: polyethylenimine-DNA complexes and time-resolved fluorescence spectroscopy, J. Phys. Chem. B, 2011, 115, 1895–1902 CrossRef CAS PubMed.
- E. Vuorimaa, A. Urtti, R. Seppanen, H. Lemmetyinen and M. Yliperttula, Time-resolved fluorescence spectroscopy reveals functional differences of cationic polymer-DNA complexes, J. Am. Chem. Soc., 2008, 130, 11695–11700 CrossRef CAS PubMed.
- T. M. Ketola, M. Hanzlikova, L. Leppanen, M. Ravina, C. J. Bishop, J. J. Green, A. Urtti, H. Lemmetyinen, M. Yliperttula and E. Vuorimaa-Laukkanen, Independent versus Cooperative Binding in Polyethylenimine-DNA and Poly(L-lysine)-DNA Polyplexes, J. Phys. Chem. B, 2013, 117, 10405–10413 CrossRef CAS PubMed.
- E. V. B. van Gaal, R. van Eijk, R. S. Oosting, R. J. Kok, W. E. Hennink, D. J. A. Crommelin and E. Mastrobattista, How to screen non-viral gene delivery systems in vitro?, J. Controlled Release, 2011, 154, 218–232 CrossRef CAS PubMed.
- G. Cosa, K. S. Focsaneanu, J. R. McLean, J. P. McNamee and J. C. Scaiano, Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution, Photochem. Photobiol., 2001, 73, 585–599 CrossRef CAS PubMed.
- W. Rettig and R. Lapouyade, Topics in fluorescence spectroscopy, Probe design and chemical sensing, in Topics in fluorescence spectroscopy, Probe design and chemical sensing, ed. J. Lakowicz, Kluwer Academic/Plenum Publishers, New York, 1994, pp. 109–149 Search PubMed.
- R. Cubeddu, D. Comelli, C. D'Andrea, P. Taroni and G. Valentini, Time-resolved fluorescence imaging in biology and medicine, J. Phys. D: Appl. Phys., 2002, 35, R61–R76 CrossRef CAS.
- K. Suhling, P. M. W. French and D. Phillips, Time-resolved fluorescence microscopy, Photochem. Photobiol. Sci., 2005, 4, 13–22 CAS.
- S. H. Huh, H. J. Do, H. Y. Lim, D. K. Kim, S. J. Choi, H. Song, N. H. Kim, J. K. Park, W. K. Chang, H. M. Chung and J. H. Kim, Optimization of 25 kDa linear polyethylenimine for efficient gene delivery, Biologicals, 2007, 35, 165–171 CrossRef CAS PubMed.
- X. B. Dou, Y. Hu, N. N. Zhao and F. J. Xu, Different types of degradable vectors from low-molecular-weight polycation-functionalized poly(aspartic acid) for efficient gene delivery, Biomaterials, 2014, 35, 3015–3026 CrossRef CAS PubMed.
- M. A. Mintzer and E. E. Simanek, Nonviral vectors for gene delivery, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed.
- R. S. Burke and S. H. Pun, Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver, Bioconjugate Chem., 2008, 19, 693–704 CrossRef CAS PubMed.
- S. Prevost, S. Riemer, W. Fischer, R. Haag, C. Bottcher, J. Gummel, I. Grillo, M. S. Appavou and M. Gradzielski, Colloidal Structure and Stability of DNA/Polycations Polyplexes Investigated by Small Angle Scattering, Biomacromolecules, 2011, 12, 4272–4282 CrossRef CAS PubMed.
- D. Pezzoli, M. Zanda, R. Chiesa and G. Candiani, The yin of exofacial protein sulfhydryls and the yang of intracellular glutathione in in vitro transfection with SS14 bioreducible lipoplexes, J. Controlled Release, 2012, 165, 44–53 CrossRef PubMed.
- C. D'Andrea, L. Spinelli, A. Bassi, A. Giusto, D. Contini, J. Swartling, A. Torricelli and R. Cubeddu, Time-resolved spectrally constrained method for the quantification of chromophore concentrations and scattering parameters in diffusing media, Opt. Express, 2006, 14, 1888–1898 CrossRef.
| This journal is © The Royal Society of Chemistry and Owner Societies 2014 |