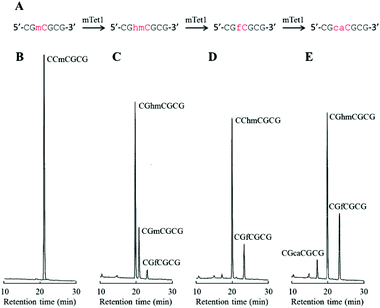
CGmCGCG is a versatile substrate with which to evaluate Tet protein activity†
Seiichiro
Kizaki
a and
Hiroshi
Sugiyama
*ab
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-Oiwakecho, Sakyo-ku, Kyoto, 606-8502, Japan. E-mail: hs@kuchem.kyoto-u.ac.jp; Fax: +81 75 753 3670; Tel: +81 75 753 4002
bInstitute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan
First published on 9th October 2013
Abstract
Tet family proteins have the ability to convert 5-methylcytosine (mC) to 5-hydroxymethylcytosine, and further to 5-formylcytosine and 5-carboxycytosine. We found that CGmCGCG can be the substrate of Tet protein, and observed iterative oxidation of mC by HPLC analysis. We also demonstrated that Tet protein favours single-stranded DNA over double-stranded DNA.
Introduction
Since the discovery of the ability of Tet (ten-eleven-translocation) protein to convert 5-methylcytosine (mC) to 5-hydroxymethylcytosine (hmC) in 2009,1 Tet family proteins have attracted a great deal of attention because of their postulated involvement in the active demethylation pathway.2,3 In 2011, it was found that Tet proteins can oxidize hmC further to 5-formylcytosine (fC) and 5-carboxycytosine (caC).4,5 Many methods to detect hmC, fC and caC in genomic DNA have been developed,6–10 and all of these oxidized derivatives of mC are known to exist in mammalian tissues.1,4,11–13To evaluate the ability of Tet proteins to oxidize mC, 20-mer or longer DNAs containing mC were incubated with Tet protein followed by enzymatic digestion, and TLC, mass spectroscopy or liquid chromatography analysis.1,4,5,14 However, these methods require many steps and are time-consuming. In this study, we present a simple and versatile method for assessment of iterative oxidation of mC by Tet protein.
Results and discussion
First, 3- to 6-mer short DNAs containing mC were synthesized using phosphoramidite chemistry. After purification, each DNA was incubated with mTet1 protein at 37 °C for 1 hour, and then the reaction mixture was directly analyzed by reversed-phase HPLC. The percentage of conversion was calculated from the peak area of the mC-containing material and the hmC-containing product (Fig. 1). Identification of the hmC-containing product was conducted by HPLC analysis following enzymatic digestion. Additionally, the reactivity of 20-mer DNAs4 was also checked by HPLC following enzymatic digestion of the product (Fig. 1 and Fig. S1, ESI†). The reactivity of short DNAs was much higher than that of 20-mer DNAs. A 6-mer DNA containing mC at a non-CpG site also reacted with mTet1 to form a hmC-containing product. For further analysis of the reactivity of short DNA, CGmCGCG was chosen as a substrate because CGmCGCG and its oxidized derivatives showed clearly distinct peaks on HPLC analysis.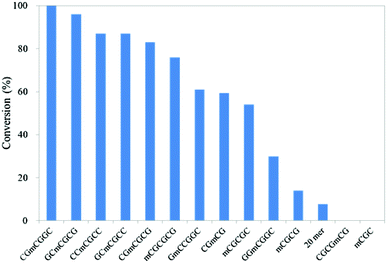 | ||
| Fig. 1 Reactivity of various DNAs with mTet1 protein. 55.4 μM of DNA and 729 nM mTet1 protein were incubated at 37 °C for 1 hour. | ||
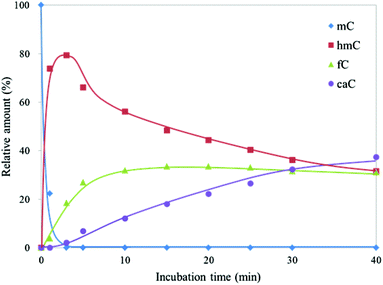
To investigate the dynamic change in the amount of CGmCGCG and its oxidized derivatives, CGmCGCG was incubated with a higher concentration of mTet1 to observe further oxidation of hmC to fC and caC. CGhmCGCG, CGfCGCG, and CGcaCGCG were synthesized using phosphoramidite chemistry and checked by reversed-phase HPLC and ESI-TOF-MS (Fig. S2 and S3, ESI†). By co-injection of these authentic samples with the reaction mixture of mTet1 and CGmCGCG, each peak was identified in two different solvent systems. After mixing CGmCGCG and mTet1, the reaction was quenched by dilution after an appropriate length of time, and then the reaction mixture was directly analyzed by HPLC (Fig. 2). The oxidation from mC to hmC was rapid and it took only about 3 minutes for CGmCGCG to be completely consumed (Fig. 3). At around 3 minutes, the amount of hmC began to decline and concomitantly the amount of caC began to gradually increase. Finally, the relative amount of each oxidized derivative reached a plateau. This may be the result of the inactivation of the mTet1 protein described in a previous report.4 Additionally, the effect of ATP in the reaction mixture on the oxidation reaction was examined by using CGmCGCG as a substrate (Fig. S4, ESI†). In the absence of ATP, the conversion efficiency from mC to hmC markedly decreased. This finding clearly shows that the activity of Tet protein is greatly regulated by ATP as described in a previous report.5
To determine the temperature dependency of the oxidation reaction of mC to hmC by mTet1 protein, CGmCGCG was incubated with mTet1 protein for 1 hour at various temperatures (Fig. 4). The optimum temperature for the oxidation reaction was around 37 °C. To our surprise, the reaction occurred even at 50 °C. At this temperature, almost all CGmCGCG strands are considered to be present as single strand, suggesting that single-stranded DNA can be the substrate of Tet protein. Therefore, the reactivity of single-stranded DNA was investigated subsequently.
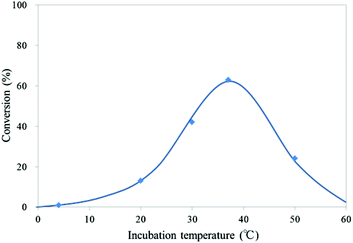 | ||
| Fig. 4 Temperature-dependency of the reaction of CGmCGCG with mTet1 protein. 55.4 μM DNA and 729 nM mTet1 protein were incubated for 1 hour. | ||
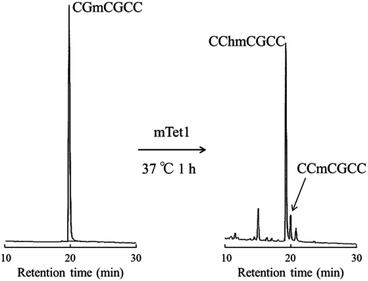
Non-self-complementary 6-mer DNA, CCmCGCC was incubated with mTet1 protein at 37 °C for 1 hour followed by direct analysis of the reaction mixture by HPLC (Fig. 5). Indeed, CCmCGCC was oxidized to form CChmCGCC, indicating that Tet protein can act on single-stranded DNA.
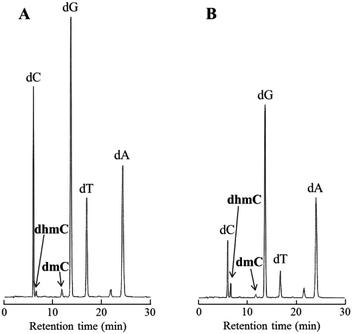
Subsequently, to examine whether Tet protein prefers mC oxidation in single-stranded or double-stranded DNA, the percentage of the conversion from mC to hmC was calculated using 20-mer DNA, 5′-GAGCGTGACmCGGAGCTGAAA-3′ in the presence or absence of its complementary strand (Fig. 6). The percentage of the conversion was 29% for double-stranded DNA, and 68% for single-stranded DNA. A similar experiment using 5′-TTTCAGCTCmCGGTCACGCTC-3′ showed the same preference (Fig. S5, ESI†). These results suggest that Tet protein has higher activity on single-stranded DNA than on double-stranded DNA.
In this study, mTet1 active domain was used for all assays. It is not clear whether full length mTet1 and the other members of Tet family proteins show similar activity. However, it was recently reported that the activity of full length mTet1 is higher than that of mTet1 active domain,5 and all Tet family proteins possess similar activity.4,5,14
Conclusions
In the present study, we demonstrated that 4- to 6-mer DNAs can be substrates of Tet protein. In particular, 6-mer DNAs were much more reactive than conventional 20-mer DNAs. Although there is a report about the effects of substrate length on AlkB, ABH2 and ABH3 proteins,15 to the best of our knowledge this is the first study which has shown the effect of substrate length on Tet protein. By using CGmCGCG, it is possible to readily observe the oxidation of mC to hmC, fC, and caC. We also showed that Tet protein can oxidize mC in single-stranded and double stranded DNA.Experimental
General
For the synthesis of 3- to 6-mer short DNAs, N4-benzoyl-5′-O-(4,4′-dimethoxytrityl)-2′-deoxycytidine-3′-O-[O-(2-cyanoethyl)-N,N′-diisopropylphosphoramidite and N2-isobutyryl-5′-O-(4,4′-dimethoxytrityl)-2′-deoxyguanosine-3′-O-[O-(2-cyanoethyl)-N,N′-diisopropylphosphoramidite were purchased from Sigma Aldrich, Japan, 5′-dimethoxytrityl-N-benzoyl-5-methyl-2′-deoxycytidine, 3′-[(2-cyanoethyl)-(N,N′-diisopropyl)]-phosphoramidite, 5′-dimethoxytrityl-N-benzoyl-5-cyanoethoxy-methyl-2′-deoxycytidine, 3′-[(2-cyanoethyl)-(N,N′-diisopropyl)]-phosphoramidite, 5′-dimethoxytrityl-N-acetyl-5-(1,2-diacetyloxy-ethyl)-2′-deoxycytidine, 3′-[(2-cyanoethyl)-(N,N′-diisopropyl)]-phosphoramidite and 5′-dimethoxytrityl-N-benzoyl-5-ethylcarboxy-2′-deoxycytidine, 3′-[(2-cyanoethyl)-(N,N′-diisopropyl)]-phosphoramidite were purchased from Glen Research. Short DNAs were synthesized on 3400 DNA synthesizer (Applied Biosystems). After purification with a high-performance liquid chromatography (HPLC) system equipped with a reversed-phase ODS column CHEMCOBOND 5-ODS-H (Chemco Scientific), synthesized short DNAs were checked by HPLC and ESI-TOF-MS Bruker BioTOF II (Bruker Daltonics). DNA concentrations were determined using Nano drop ND-1000 (Thermo Scientific) (50 cm−1 M−1). The sequences of 20-mer DNAs are 5′-TTTCAGCTCmCGGTCACGCTC-3′, 5′-GAGCGTGACmCGGAGCTGAAA-3′, 5′-TTTCAGCTCCGGTCACGCTC-3′ and 5′-GAGCGTGACCGGAGCTGAAA-3′. These 20-mer DNAs were purchased from Japan Bio Services (for mC-containing DNA) and Sigma Aldrich Japan.Tet activity assay
mTet1 active domain (1367–2039) was purchased from Wisegene, stocked in 20 mM HEPES (pH 7.4), NaCl 50 mM, glycerol 50%. DNAs containing mC were incubated with mTet1 protein in 50 mM HEPES (pH 8.0), 100 mM NaCl, 2 mM L-ascorbic acid, 1 mM 2-oxoglutarate disodium salt hydrate, 105 μM Fe(NH4)2(SO4)2 6H2O, 1.2 mM ATP and 2.5 mM DTT.For analysis of the reactivity of 3- to 6-mer DNAs with mTet1 protein, 55.4 μM DNA and 729 nM mTet1 protein were incubated at 37 °C for 1 hour (total volume: 25 μL). After incubation, 3 μL of the reaction mixture was used for HPLC analysis. For analysis of the reactivity of 20-mer DNAs with mTet1 protein, 55.4 μM DNA and 729 nM mTet1 protein were incubated at 37 °C for 1 hour (total volume: 25 μL). After incubation, the reaction mixture was purified using QIAquick Nucleotide Removal Kit (Qiagen) followed by incubation with nuclease P1 (Wako) and Antarctic Phosphatase (New England Biolabs) at 37 °C for 4 hours. All of the reaction mixture was used for HPLC analysis. For time-course analysis of the reaction of CGmCGCG with mTet1 protein, 55.4 μM DNA and 7.29 μM mTet1 protein were incubated at 37 °C for 1, 3, 5, 10, 15, 25, 30, 40 minutes (total volume: 25 μL). After incubation, the reaction mixture was quenched by dilution, and 3 μL of the reaction mixture was used for HPLC analysis. For temperature-dependency analysis of the reaction of CGmCGCG with mTet1 protein, 55.4 μM DNA and 729 nM mTet1 protein were incubated at 4, 20, 30, 37, 50 °C for 1 hour (total volume: 25 μL). After incubation, 3 μL of the reaction mixture was used for HPLC analysis. For analysis of the reactivity of CCmCGCC with mTet1 protein, 55.4 μM DNA and 729 nM mTet1 protein were incubated at 37 °C for 1 hour (total volume: 25 μL). After incubation, 3 μL of the reaction mixture was used for HPLC analysis.
For analysis of the reactivity of 20-mer DNA with mTet1 protein in the absence or presence of its complementary strand, 16.3 μM DNA and 729 nM mTet1 protein were incubated at 37 °C for 1 hour (total volume: 50 μL). After incubation, all of the reaction mixture was purified using QIAquick Nucleotide Removal Kit (Qiagen). The purified DNAs were digested with nuclease P1 (Wako) and Antarctic phosphatase (New England Biolabs) at 37 °C for 4 hours. All of the reaction mixture was used for HPLC analysis. Elution was done with 50 mM ammonium formate containing 0–3% acetonitrile in a linear gradient at a flow rate of 1.0 mL min−1 for 30 minutes, at 40 °C.
Notes and references
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930 CrossRef CAS PubMed.
- J. U. Guo, Y. Su, C. Zhong, G. Ming and H. Song, Cell, 2011, 145, 423 CrossRef CAS PubMed.
- S. C. Wu and Y. Zhang, Nat. Rev. Mol. Cell Biol., 2010, 11, 607 CrossRef CAS PubMed.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300 CrossRef CAS PubMed.
- Y.-F. He, B.-Z. Li, Z. Li, P. Liu, Y. Wang, Q. Tang, J. Ding, Y. Jia, Z. Chen, L. Li, Y. Sun, X. Li, Q. Dai, C.-X. Song, K. Zhang, C. He and G.-L. Xu, Science, 2011, 333, 1303 CrossRef CAS PubMed.
- M. Yu, G. C. Hon, K. E. Szulwach, C.-X. Song, L. Zhang, A. Kim, X. Li, Q. Dai, Y. Shen, B. Park, J. Min, P. Jin, B. Ren and C. He, Cell, 2012, 149, 1368 CrossRef CAS PubMed.
- M. J. Booth, M. R. Branco, G. Ficz, D. Oxley, F. Krueger, W. Reik and S. Balasubramanian, Science, 2012, 336, 934 CrossRef CAS PubMed.
- P. Schüler and A. K. Miller, Angew. Chem., Int. Ed., 2012, 51, 10704 CrossRef PubMed.
- C.-X. Song, K. E. Szulwach, Q. Dai, Y. Fu, S.-Q. Mao, L. Lin, C. Street, Y. Li, M. Poidevin, H. Wu, J. Gao, P. Liu, L. Li, G.-L. Xu, P. Jin and C. He, Cell, 2013, 153, 1 Search PubMed.
- X. Lu, C. Song, K. Szulwach, Z. Wang, P. Weidenbacher, P. Jin and C. He, J. Am. Chem. Soc., 2013, 135, 9315 CrossRef CAS PubMed.
- S. Kriaucionis and N. Heintz, Science, 2009, 324, 929 CrossRef CAS PubMed.
- M. Münzel, D. Globisch, T. Brückl, M. Wagner, V. Welzmiller, S. Michalakis, M. Müller, M. Biel and T. Carell, Angew. Chem., Int. Ed., 2010, 49, 5375 CrossRef PubMed.
- T. Pfaffeneder, B. Hackner, M. Truß, M. Münzel, M. Müller, C. A. Deiml, C. Hagemeier and T. Carell, Angew. Chem., Int. Ed., 2011, 123, 7146 CrossRef.
- S. Ito, A. C. D'Alessio, O. V. Taranova, K. Hong, L. C. Sowers and Y. Zhang, Nature, 2010, 466, 1129 CrossRef CAS PubMed.
- P. Koivisto, T. Duncan, T. Lindahl and B. Sedgwick, J. Biol. Chem., 2003, 278, 44348 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–4. See DOI: 10.1039/c3ob41823e |
| This journal is © The Royal Society of Chemistry 2014 |