Protein coronas suppress the hemolytic activity of hydrophilic and hydrophobic nanoparticles†
Krishnendu
Saha‡
,
Daniel F.
Moyano‡
and
Vincent M.
Rotello
*
Department of Chemistry, University of Massachusetts Amherst, 710 North Pleasant St, Amherst, Massachusetts, USA 01003. E-mail: rotello@chem.umass.edu; Fax: +413-545-4490; Tel: +413-545-2058
First published on 27th September 2013
Abstract
The role of nanoparticle surface hydrophobicity on hemolytic properties is established in the absence and presence of plasma proteins. Significantly, the formation of a plasma protein corona on the NP surface protects red blood cells from both hydrophilic and hydrophobic NP-mediated hemolysis.
Intravascular rupture of red blood cells (RBCs) upon systemic administration of therapeutic NPs can elevate the plasma level of cell-free hemoglobin leading to endothelial cell dysfunction and vascular thrombosis.1 This acute hemolysis can further cause hemolytic anemia that results in multiple organ failure2 and death.3 Several reports have recently demonstrated the hemolytic activities of therapeutic NPs both in vitro4 and in vivo,5 indicating potential adverse side-effects of NPs that can limit applications in nanomedicine. However, a lack of parametric study in vitro of NP hemolytic behavior makes their interaction profile difficult to predict in vivo.
Surface functionality of NPs is central to their effective use in therapeutic applications, imparting functional properties6 and dictating their circulation profile in the blood stream.7 However, in contrast to small molecule therapeutics the formation of a protein corona on the NP surface during blood circulation8 alters the surface chemical behavior of NPs and defines the physiological responses.9 This interplay between the NP surface functionality and the protein corona formed in situ would be expected to dictate the overall interaction of NPs with RBCs, regulating the hemolytic behavior of NPs. To date, however, there is no clear understanding of how changes on the NP surfaces coupled with the formation of a protein corona control the hemolytic properties of NPs.10
Herein, we report on the role of NP surface functionality on hemolytic activity in the presence and absence of plasma proteins. To this end, we have synthesized a class of cationic gold NPs of the same core size (∼2 nm), differing in their surface hydrophobicity (Fig. 1). These NPs showed a direct increase in hemolytic activity with an increase of the surface hydrophobicity in the absence of plasma proteins. Significantly, the presence of a protein corona dramatically altered the hemolytic activity of these NPs; hemolysis was only observed for the most hydrophobic surface coverage. These studies demonstrate the importance of plasma proteins in moderating hemolytic activity, expanding the range of particle surfaces that can be employed without hemolytic consequences.
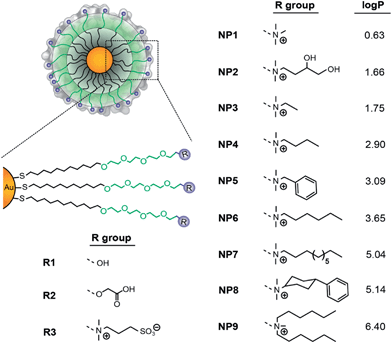 | ||
Fig. 1 The structure of the NPs used in the current study; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P denotes the n-octanol/water partition coefficients of the R groups. P denotes the n-octanol/water partition coefficients of the R groups. | ||
The NP surface hydrophobicity has a critical role in the cellular uptake,11 toxicity,12 and immune responses13 of nanomaterials. To understand the role of NP surface hydrophobicity in blood compatibility, a family of nine cationic NPs was synthesized systematically changing the degree of surface hydrophobicity by the use of specific chemical groups (Fig. 1).14 This chemical approach allows us to control the nature of the monolayer (nanoparticle surface), while keeping the other physico-chemical properties (e.g. size and surface charge) constant. Using this system, the NP properties can be translated into numerical descriptors, as demonstrated previously by interfacial tension experiments.15 As such, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of the headgroups (R groups, Fig. 1) were employed to describe the relative hydrophobic nature of the NP surfaces.
P values of the headgroups (R groups, Fig. 1) were employed to describe the relative hydrophobic nature of the NP surfaces.
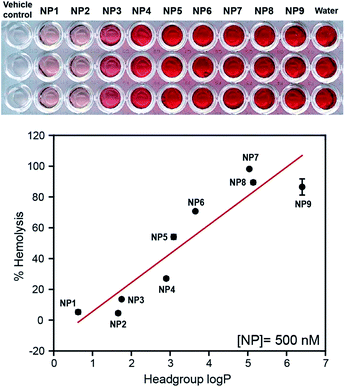
In previous studies, we have demonstrated that cationic NPs showed significantly higher hemolysis compared to anionic counterparts.16 To this end, NPs with different surface charges, e.g. cationic, anionic, neutral, and zwitterionic (500 nM each) were incubated with RBCs for 30 min and the absorbance of released hemoglobin from the hemolyzed RBCs was measured at 570 nm.17 We observed a significantly higher hemolytic activity for cationic NPs compared to NPs with other surface charges (anionic, neutral, and zwitterionic) (see ESI†), corroborating previous reports.18
The role of NP surface hydrophobicity on hemolytic behavior was established by incubating NP1–NP9 (500 nM each) with RBCs as mentioned above. A direct increase in hemolytic activity was observed for NP1–NP9 with an increase of the hydrophobicity (Fig. 2), demonstrating the role of the NP surface hydrophobicity on the hemolysis. Significantly, NPs with similar headgroup log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values but different chemical functionalities, e.g.NP2 and NP3, demonstrated different degrees of hemolysis (∼4.5% and 13.5%, respectively), providing evidence for the role of specific functional groups in the observed hemolytic behavior.
P values but different chemical functionalities, e.g.NP2 and NP3, demonstrated different degrees of hemolysis (∼4.5% and 13.5%, respectively), providing evidence for the role of specific functional groups in the observed hemolytic behavior.
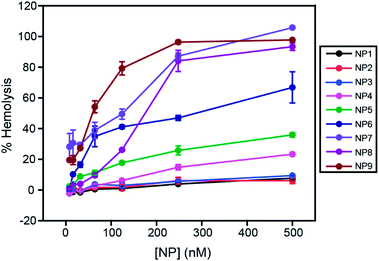
The dose-dependent hemolytic behavior of these NPs was investigated by exposing RBCs to a range of concentrations of NPs from 8 nM to 500 nM. As shown in Fig. 3, a dose-dependent increase of hemolysis was observed for all NPs. NP1–NP3 did not show significant hemolysis at the highest concentration tested (500 nM), demonstrating biocompatibility of these particles with RBCs. Significantly, a sigmoidal dose–response curve was observed for more hydrophobic NPs (see ESI†), demonstrating the co-operative nature of the hemolytic process for these NPs.19 This result demonstrated that the subtle changes on NP surfaces can modulate the interaction profile with RBCs, leading to different hemolytic profiles.
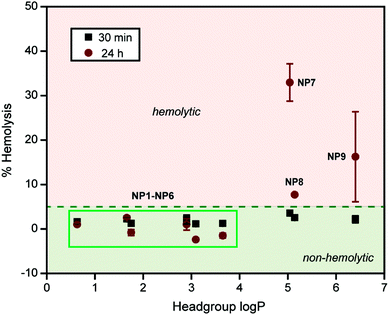
The binding of plasma proteins on a NP can mask the chemical nature of the NP surface, altering its activity in vivo.20 The hemolytic activities of NP1–NP9 were further studied in the presence of 55% plasma protein, a condition NPs will meet when administered in the blood stream. NPs were pre-incubated with 55% plasma in PBS for 30 min and then added to the RBCs. The pre-incubation time was chosen to ensure protein absorption, although protein coronas form within minutes after NP exposure.21 Following incubation with RBCs, the absorbance of the supernatant was measured at 570 nm to monitor the extent of hemolysis (vide supra). In presence of plasma, little to no hemolysis was observed for NP1–NP6 (Fig. 4). Significantly, NP6, which showed ∼70% hemolysis in the absence of plasma protein, demonstrated no hemolytic activity in the presence of plasma even after 24 h, a striking example of the effect of protein corona formation on the NP surface. Likewise, NP7–NP9 demonstrated a significant decrease in hemolytic activity (more than 20-fold) in the presence of plasma within 30 minutes. However, NP7–NP9 still showed ∼5% hemolysis within 30 min, demonstrating the hemolytic potential of these NPs even in the presence of plasma proteins. This behavior was more pronounced after 24 h where NP7 and NP9 showed severe hemolysis (Fig. 4). Significantly, NP7 and NP8 showed different hemolytic activities after 24 h in the presence of plasma proteins, despite their similar headgroup hydrophobicity, suggesting that surface topology plays a role in hemolysis propensity. Nonetheless, this study signifies the interplay of the chemical functionalities on the NP surface and the protein corona formed in situ, making NPs ‘silent’ towards hemolytic consequences. However, NPs with a high degree of surface hydrophobicity (headgroup log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 4) demonstrated severe hemolysis, providing a limit to the NP surface hydrophobicities tested in this study.
P > 4) demonstrated severe hemolysis, providing a limit to the NP surface hydrophobicities tested in this study.
Conclusion
In summary, we have demonstrated the important synergy between the NP surface chemical functionalities and the protein corona, leading to dramatically attenuated hemolytic behaviour of NPs. In the absence of plasma proteins, a linear hemolytic profile was observed with increasing NP surface hydrophobicity. Significantly, the presence of plasma protein passivated both hydrophilic and hydrophobic NP surfaces leading to no hemolysis within 30 min. NPs with the highest degrees of hydrophobicity (headgroup log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P > 4), however, maintained their hemolytic properties (for 24 h), potentially due to aggregation in the presence of plasma proteins. This study demonstrates the protective role of the protein corona for improved blood compatibility and provides extended design parameters for therapeutic nanomaterials without toxic consequences.
P > 4), however, maintained their hemolytic properties (for 24 h), potentially due to aggregation in the presence of plasma proteins. This study demonstrates the protective role of the protein corona for improved blood compatibility and provides extended design parameters for therapeutic nanomaterials without toxic consequences.
Acknowledgements
This research was supported in part by a grant from the National Institute of Health (grants EB014277). NSF support is acknowledged for NSEC Center for Hierarchical Manufacturing (CMMI-1025020, BY) and MRSEC facilities (DMR-0820506).Notes and references
- R. P. Rother, L. Bell, P. Hillmen and M. T. Gladwin, JAMA, J. Am. Med. Assoc., 2005, 293, 1653–1662 CrossRef CAS PubMed.
- (a) D. Stroncek, J. L. Procter and J. Johnson, Am. J. Hematol., 2000, 64, 67–70 CrossRef CAS; (b) D. Saitoh, T. Kadota, A. Senoh, T. Takahara, Y. Okada, K. Mimura, H. Yamashita, H. Ohno and M. Inoue, Am. J. Emerg. Med., 1993, 11, 355–359 CrossRef CAS; (c) M. Scharte and M. P. Fink, Crit. Care Med., 2003, 31, S651–S657 CrossRef CAS PubMed.
- (a) J. Perkins, Asian J. Transfus. Sci., 2008, 2, 20–23 CrossRef PubMed; (b) S. Yuerek, H. Riess, S. Kreher, B. Doerken and A. Salama, Transfus. Med., 2010, 20, 265–268 CrossRef PubMed.
- (a) S. A. Love, J. W. Thompson and C. L. Haynes, Nanomedicine, 2012, 7, 1355–1364 CrossRef CAS PubMed; (b) J. Choi, V. Reipa, V. M. Hitchins, P. L. Goering and R. A. Malinauskas, Toxicol. Sci., 2011, 123, 133–143 CrossRef CAS PubMed; (c) Y. N. Zhao, X. X. Sun, G. N. Zhang, B. G. Trewyn, I. I. Slowing and V. S. Y. Lin, ACS Nano, 2011, 5, 1366–1375 CrossRef CAS PubMed.
- (a) S. L. Lu, R. Duffin, C. Poland, P. Daly, F. Murphy, E. Drost, W. MacNee, V. Stone and K. Donaldson, Environ. Health Perspect., 2009, 117, 241–247 CAS; (b) Y. T. Li, J. Liu, Y. J. Zhong, J. Zhang, Z. Y. Wang, L. Wang, Y. L. An, M. Lin, Z. Q. Gao and D. S. Zhang, Int. J. Nanomed., 2011, 6, 2805–2819 CrossRef CAS PubMed.
- (a) K. Saha, A. Bajaj, B. Duncan and V. M. Rotello, Small, 2011, 7, 1903–1918 CrossRef CAS PubMed; (b) R. Mout, D. F. Moyano, S. Rana and V. M. Rotello, Chem. Soc. Rev., 2012, 41, 2539–2544 RSC.
- J. W. Yoo, E. Chambers and S. Mitragotri, Curr. Pharm. Des., 2010, 16, 2298–2307 CrossRef CAS.
- (a) C. D. Walkey and W. C. W. Chan, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC; (b) M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed; (c) R. R. Arvizo, K. Giri, D. Moyano, O. R. Miranda, B. Madden, D. J. McCormick, R. Bhattacharya, V. M. Rotello, J. P. Kocher and P. Mukherjee, PLoS One, 2012, 7, e33650 CAS.
- (a) C. Salvador-Morales, E. Flahaut, E. Sim, J. Sloan, M. L. H. Green and R. B. Sim, Mol. Immunol., 2006, 43, 193–201 CrossRef CAS PubMed; (b) D. E. Owens and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef CAS PubMed.
- (a) M. Joglekar, R. A. Roggers, Y. N. Zhao and B. G. Trewyn, RSC Adv., 2013, 3, 2454–2461 RSC; (b) T. Kwon, H. J. Woo, Y. H. Kim, H. J. Lee, K. H. Park, S. Park and B. Youn, J. Nanosci. Nanotechnol., 2012, 12, 6168–6175 CrossRef CAS PubMed.
- Z. J. Zhu, T. Posati, D. F. Moyano, R. Tang, B. Yan, R. W. Vachet and V. M. Rotello, Small, 2012, 8, 2659–2663 CrossRef CAS PubMed.
- A. Chompoosor, K. Saha, P. S. Ghosh, D. J. Macarthy, O. R. Miranda, Z. J. Zhu, K. F. Arcaro and V. M. Rotello, Small, 2010, 6, 2246–2249 CrossRef CAS PubMed.
- D. F. Moyano, M. Goldsmith, D. J. Solfiell, D. Landesman-Milo, O. R. Miranda, D. Peer and V. M. Rotello, J. Am. Chem. Soc., 2012, 134, 3965–3967 CrossRef CAS PubMed.
- D. F. Moyano and V. M. Rotello, Langmuir, 2011, 27, 10376–10385 CrossRef CAS PubMed.
- S. Rana, X. Yu, D. Patra, D. F. Moyano, O. R. Miranda, I. Hussain and V. M. Rotello, Langmuir, 2012, 28, 2023–2027 CrossRef CAS PubMed.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- (a) Y. S. Lin and C. L. Haynes, Chem. Mater., 2009, 21, 3979–3986 CrossRef CAS; (b) Y. S. Lin and C. L. Haynes, J. Am. Chem. Soc., 2010, 132, 4834–4842 CrossRef CAS PubMed; (c) T. Yu, A. Malugin and H. Ghandehari, ACS Nano, 2011, 5, 5717–5728 CrossRef CAS PubMed.
- (a) R. R. Arvizo, O. R. Miranda, M. A. Thompson, C. M. Pabelick, R. Bhattacharya, J. D. Robertson, V. M. Rotello, Y. S. Prakash and P. Mukherjee, Nano Lett., 2010, 10, 2543–2548 CrossRef CAS PubMed; (b) N. M. Schaeublin, L. K. Braydich-Stolle, A. M. Schrand, J. M. Miller, J. Hutchison, J. J. Schlager and S. M. Hussain, Nanoscale, 2011, 3, 410–420 RSC; (c) P. R. Leroueil, S. A. Berry, K. Duthie, G. Han, V. M. Rotello, D. Q. McNerny, J. R. Baker, B. G. Orr and M. M. B. Holl, Nano Lett., 2008, 8, 420–424 CrossRef CAS PubMed.
- Z. I. Andreeva, V. F. Nesterenko, M. G. Fomkina, V. I. Ternovsky, N. E. Suzina, A. Y. Bakulina, A. S. Solonin and E. V. Sineva, Biochim. Biophys. Acta, 2007, 1768, 253–263 CrossRef CAS PubMed.
- (a) I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS; (b) M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS PubMed.
- (a) D. Dell'Orco, M. Lundqvist, C. Oslakovic, T. Cedervall and S. Linse, PLoS One, 2010, 5, e10949 Search PubMed; (b) A. L. Barran-Berdon, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Lagana, Langmuir, 29, 6485–6494 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: NP synthesis, hydrodynamic sizes, zeta potentials, and pictures of NP hemolysis. See DOI: 10.1039/c3mh00075c |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2014 |