Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High-content screening of yeast mutant libraries by shotgun lipidomics†
Kirill
Tarasov
*ab,
Adam
Stefanko
cd,
Albert
Casanovas
c,
Michal A.
Surma
d,
Zane
Berzina
c,
Hans Kristian
Hannibal-Bach
c,
Kim
Ekroos
a and
Christer S.
Ejsing
*c
aZora Biosciences Oy, Biologinkuja 1, FI-02150 Espoo, Finland. E-mail: kirill.tarasov@outlook.com
bDepartment of Biochemistry and Molecular Medicine, Université de Montréal, H3T 1J4, Montréal, Canada
cDepartment of Biochemistry and Molecular Biology, University of Southern Denmark, 5230 Odense, Denmark. E-mail: cse@bmb.sdu.dk
dMax Planck Institute of Molecular Cell Biology and Genetics, 01307 Dresden, Germany
First published on 21st March 2014
Abstract
To identify proteins with a functional role in lipid metabolism and homeostasis we designed a high-throughput platform for high-content lipidomic screening of yeast mutant libraries. To this end, we combined culturing and lipid extraction in 96-well format, automated direct infusion nanoelectrospray ionization, high-resolution Orbitrap mass spectrometry, and a dedicated data processing framework to support lipid phenotyping across hundreds of Saccharomyces cerevisiae mutants. Our novel approach revealed that the absence of genes with unknown function YBR141C and YJR015W, and the transcription factor KAR4 precipitated distinct lipid metabolic phenotypes. These results demonstrate that the high-throughput shotgun lipidomics platform is a valid and complementary proxy for high-content screening of yeast mutant libraries.
Introduction
The lipidome of eukaryotic cells consists of several hundreds to thousands of molecular lipid species that constitute membranes, store metabolic energy and function as signalling molecules.1,2 The structural heterogeneity of lipids is defined by a metabolic network of enzymes and regulatory factors that synthesize distinct lipid species by assembling or disassembling a multitude of available hydrocarbon residues and polar head groups. Lipid species can be divided into several categories based on their chemical structures.3 The most abundant lipid categories in eukaryotic cells include glycerophospholipids, sphingolipids, glycerolipids and sterol lipids, which mediate distinct molecular functions. Notably, several metabolic transitions interlink glycerophospholipid, sphingolipid, glycerolipid and sterol metabolism such that perturbations are prone to induce lipidome-wide ripple effects and prompt compensatory responses to sustain lipid homeostasis.4 Compromising the lipid metabolic network is known to cause dysfunctional lipid homeostasis and cellular lipotoxicity that precipitate disorders such as obesity, atherosclerosis and neurodegeneration.5 Importantly, the regulatory mechanisms that govern global lipid metabolism and relay physiological signals to sustain lipid homeostasis are largely unknown.Genetic and biochemical studies using the yeast Saccharomyces cerevisiae have been instrumental in elucidating the blueprint of lipid metabolism and defining the physiological functions of lipids.6,7 Early efforts have pinned lipid metabolism to the framework of global metabolism as illustrated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway resource.8 Moreover, genetic and biochemical approaches combined with molecular biology have paved the way to cloning and functional characterization of key enzymes in the human lipid metabolic network.6 The functional regulation of lipid metabolism depends in part on a transcriptional circuitry that sets the cellular concentration of lipid enzymes and accessory regulatory factors depending on physiological requirements. The circuitry in S. cerevisiae includes the transcriptional regulators Opi1p, Ino2p, Ino4p and Zap1p that control the expression level of a subset of proteins required for glycerophospholipid metabolism.9 In addition, the circuitry also includes the regulators Mga2p and Spt23p, which are involved in controlling the expression of the fatty acid desaturase OLE1.10 Notably, regulatory mechanisms controlling most of enzymes in the lipid metabolic network are still poorly understood. Intriguingly, this raises the question of how the expression levels of enzymes involved in, for example, sphingolipid and sterol lipid metabolism are controlled.
Functional genomics strategies including gene–gene11,12 and protein–protein interactions assays13–15 can be powerful approaches for identifying regulatory factors in lipid metabolism. Recently, epistatic miniarray profiling has been instrumental in defining the molecular mechanisms of how the fatty acid chain length is determined by conserved membrane-imbedded elongase complexes,16 how conserved Orm proteins interact with the serine palmitoyltransferase to control sphingolipid biosynthesis17 and linking the GDP/GTP exchange factor Rom2p to the regulation of sphingolipid metabolism.18 These findings were prompted by high-content datasets showing particular genes interacting with known constituents of the lipid metabolic network. Notably, such interactions can also be observed in other types of publically available resource data. For example, the yeast protein interactome displays an overrepresentation of interactions between genes involved in lipid metabolism and genes with other cellular functions.13 Thus, public repositories of gene and protein interaction data combined with function prediction algorithms can be a potential resource for shortlisting genes/proteins with a putative functional role in lipid metabolism.19 Importantly, several studies have successfully combined function prediction methods with experimental confirmation to elucidate the molecular mechanisms of mitochondrial biogenesis in yeast20 and tissue-specific regulation patterns in worm.21 The success of these studies demonstrates that mining resource data and integrating lipidomic analysis can be an avenue for identifying novel lipid enzyme activities and regulators of global lipid metabolism.
Shotgun lipidomics is a relatively novel omics tool that affords comprehensive and quantitative profiling of cellular lipids. The efficacy of the technology has been documented in numerous studies of biological membrane organization, lipid–protein interactions and the regulation of lipid metabolism.17,22–25 Shotgun lipidomics implies that lipid extracts of cells are directly infused into a mass spectrometer without up-front time-consuming liquid chromatographic separation thereby shortening the time required for analysis, and that identification of lipid species relies on accurately determined masses and/or tandem mass spectra acquired from corresponding lipid species.1,26 Shotgun lipidomics enables extensive lipidome characterization by combining analyses of the same lipid extract in positive and negative ion mode, and by implementing data processing routines to merge, normalize and visualize lipidomic datasets.27 In addition, a more recent shotgun lipidomics technology based on high-resolution Orbitrap mass spectrometry and automated direct infusion nanoelectrospray ionization offers high-throughput capabilities with high sensitivity, a broad dynamic quantification range and extensive lipidome coverage spanning lipid species molar abundances over 3 to 4 orders of magnitude.4,28,29 Notably, these analytical hallmarks are ideally suited for exploratory lipidome analysis in yeast and provide a mean to screen libraries of mutant strains to identify regulatory modules in global lipid metabolism.
Here we describe a high-throughput platform for high-content lipidomic screening of yeast mutant libraries that utilizes culturing and lipid extraction in 96-well format, automated direct infusion nanoelectrospray ionization, high-mass resolution Orbitrap mass spectrometry and a dedicated data processing framework to support systematic monitoring of lipid species across hundreds of yeast strains. To catalog lipid phenotypes, we made use of ‘robust principal component analysis’ and a quantitative scoring system that we term SoamD (sum of absolute mol% difference). As a test bed, we employed the platform to array a shortlist of deletion mutants of distinct transcriptional regulators and genes with unknown function predicted to play a role in lipid metabolism. Our novel approach revealed that the absence of genes with previously unknown function YBR141C and YJR015W, and the transcription factor KAR4 precipitates distinct lipid metabolic phenotypes. These results show that combining functional genomic workflows and high-content lipidomic profiling can be a powerful proxy for identifying regulators of global lipid metabolism.
Materials and methods
Chemicals and lipid standards
Synthetic lipid standards were purchased from Avanti Polar Lipids and Larodan Fine Chemicals. Chemicals, growth media and solvents were purchased from Sigma-Aldrich, Rathburn Chemicals, MP Biomedicals and BD Biosciences.Yeast strains
In this study we used S. cerevisiae reference strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and the congenic deletion mutants listed in Table S1 (ESI†). All strains were obtained from EUROSCARF. Mutant strains without genes encoding transcriptional regulators were shortlisted based on gene ontology (GO) annotation in the Saccharomyces Genome Database (SGD).30Prediction of uncharacterized genes with potential function in lipid metabolism
A list of genes known to be implicated in lipid metabolism (query list of lipid-related genes) was compiled based on automated extraction of gene names with GO annotation ‘lipid metabolism’ or ‘lipid binding’ in the SGD, and thorough manual annotation based on literature (Table S2, ESI†). To predict uncharacterized genes with potential function in lipid metabolism we used the query list of lipid-related genes and the GeneMANIA function prediction algorithm.31 Based on available protein–protein interaction data from BioGRID,32 the GeneMANIA function prediction algorithm was requested to output the 50 most related genes to the genes on the lipid-related query list (Table S3, ESI†). From the 50 top scoring genes we selected 8 genes that were annotated with “biological process unknown”, and shortlisted the corresponding deletion mutants for first round lipidomic screening.First round screening: 96-well plate culturing
Yeast strains were plated and cultured at 30 °C for 24 hours in 0.3 ml 96-well plates (Eppendorf AG) on an agar-based solid synthetic complete medium containing 2% glucose and supplemented with 100 μM inositol and 100 μM choline. In addition to shortlisted deletion mutants each 96-well plate contained three replicates of the control strains BY4742 and elo2Δ. Yeast cells were harvested by resuspension in 100 μl of 155 mM ammonium acetate (average yield 1.5–2.0 OD600 units), transferred to 2 ml 96-well plates (Eppendorf AG) and stored at −80 °C until lipid extraction.Lipid extraction in 96-well plates at 4 °C
The yeast cell suspensions (100 μl) in 2 ml 96-well plates were added to glass beads (425–600 μm, Sigma-Aldrich) and subjected to cell disruption for 120 min at 1400 rpm and 4 °C on a ThermoMixer (Eppendorf AG). (This high-throughput oriented cell lysis procedure was benchmarked against conventional glass bead lysis4 by showing no differences in lipid profile (data not shown).) Cell lysates (70 μl) were transferred into a new 2 ml polytetrafluoroethylene 96-well plate (Radleys Discovery Technologies) and subjected to single-step lipid extraction in the 96-well plate. Samples were extracted by adding 250 μl of chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and mixing in a ThermoMixer for 120 min. The lower organic phase was collected by transferring 47 μl into two separate 150 μl 96-well plates (Eppendorf AG) that were subsequently subjected to vacuum evaporation.
1, v/v) and mixing in a ThermoMixer for 120 min. The lower organic phase was collected by transferring 47 μl into two separate 150 μl 96-well plates (Eppendorf AG) that were subsequently subjected to vacuum evaporation.
Mass spectrometric lipid analysis and data processing for 96-well plate cultures
Lipid extracts in 96-well plates were dissolved in 20 μl of 7.5 mM ammonium acetate in chloroform/methanol/propanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, V/V/V) for positive ion mode mass analysis, and 20 μl of 0.0075% methylamine in methanol/chloroform (1
4, V/V/V) for positive ion mode mass analysis, and 20 μl of 0.0075% methylamine in methanol/chloroform (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, V/V) for negative ion mode analysis. The 96-well plates were covered with aluminum sealing tape to avoid sample evaporation. Samples were analyzed by direct infusion on a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a robotic TriVersa NanoMate ion source (Advion Biosciences) as previously described.4,29 Positive ion mode analysis was performed using multiplexed FT MS with scan ranges m/z 220–530 (for monitoring lysophosphatidylcholine (LPC) and lysophosphatidylethanolamine (LPE) species) and m/z 500–1200 (for monitoring phosphatidylcholine (PC), phosphatidylethanolamine (PE), diacylglycerol (DAG), triacylglycerol (TAG), sterol ester (SE) and ceramide (Cer) species). Negative ion mode analysis was performed using multiplexed FT MS with scan ranges m/z 200–605 (for monitoring lysophosphatidic acid (LPA), lysophosphatidylserine (LPS) and lysophosphatidylinositol (LPI) species) and m/z 505–1400 (for monitoring phosphatidylinositol (PI), phosphatidic acid (PA), phosphatidylserine (PS), and inositol-phosphoceramide (IPC) species). The total time of FT MS analysis was 3 min per polarity per sample. All FT MS spectra were acquired in profile mode using a target mass resolution of 100
5, V/V) for negative ion mode analysis. The 96-well plates were covered with aluminum sealing tape to avoid sample evaporation. Samples were analyzed by direct infusion on a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a robotic TriVersa NanoMate ion source (Advion Biosciences) as previously described.4,29 Positive ion mode analysis was performed using multiplexed FT MS with scan ranges m/z 220–530 (for monitoring lysophosphatidylcholine (LPC) and lysophosphatidylethanolamine (LPE) species) and m/z 500–1200 (for monitoring phosphatidylcholine (PC), phosphatidylethanolamine (PE), diacylglycerol (DAG), triacylglycerol (TAG), sterol ester (SE) and ceramide (Cer) species). Negative ion mode analysis was performed using multiplexed FT MS with scan ranges m/z 200–605 (for monitoring lysophosphatidic acid (LPA), lysophosphatidylserine (LPS) and lysophosphatidylinositol (LPI) species) and m/z 505–1400 (for monitoring phosphatidylinositol (PI), phosphatidic acid (PA), phosphatidylserine (PS), and inositol-phosphoceramide (IPC) species). The total time of FT MS analysis was 3 min per polarity per sample. All FT MS spectra were acquired in profile mode using a target mass resolution of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, isolation waveforms enabled, automatic gain control at 1e6, max injection time at 250 ms and acquisition of 2 μscans. Lipid species were identified, quantified and visualized using ALEX software,27 SAS software (SAS Institute Inc.) and Tableau Desktop software (Tableau Software), respectively. Lipid species were annotated according to their sum composition.29 Lipid species abundance was monitored by intensity profiling using the proxy intensity% (I%) calculated as the intensity of a given lipid species divided by the sum of intensities of all monitored lipid species in a given ion mode (i.e. positive or negative). A quality control procedure having two filters was implemented: (i) strains with less than 70% of the average number of detected lipid species in at least one ion mode were rejected, (ii) strains with less than 15% of the average total lipid intensity in at least one ion mode were rejected.
000, isolation waveforms enabled, automatic gain control at 1e6, max injection time at 250 ms and acquisition of 2 μscans. Lipid species were identified, quantified and visualized using ALEX software,27 SAS software (SAS Institute Inc.) and Tableau Desktop software (Tableau Software), respectively. Lipid species were annotated according to their sum composition.29 Lipid species abundance was monitored by intensity profiling using the proxy intensity% (I%) calculated as the intensity of a given lipid species divided by the sum of intensities of all monitored lipid species in a given ion mode (i.e. positive or negative). A quality control procedure having two filters was implemented: (i) strains with less than 70% of the average number of detected lipid species in at least one ion mode were rejected, (ii) strains with less than 15% of the average total lipid intensity in at least one ion mode were rejected.
Classification of mutant strains into growth phase categories
The lipidomes of mutant strains surviving the quality control procedure were classified according to the growth phase. To this end, the BY4742 reference strain was cultured in liquid medium as described below in the section ‘Second round screening: liquid culturing’. Samples were collected for BY4742 cells in the exponential phase (0.8–3.2 OD600 units per ml) and the stationary phase (4–4.5 OD600 units per ml) determined using a growth curve. These samples were subjected to two-step lipid extraction as described below, and lipidomic analysis using intensity profiling I% as described in the previous section. To classify mutant strains according to the growth phase we performed average linkage agglomerative hierarchical clustering on I% values of lipid species and lipid classes that were the most different between the growth phases (i.e. sum of I% of lipid species belonging to TAG, PC, DAG, PI, PS, PE and PA lipid classes, and the sum of I% of PC, PI, PS, PE, PA and DAG species with carbon index up to 32, and equal or greater than 34). Analysis was performed using Cluster 3.0 software.33 Results were visualized as a clustering heatmap using I% standardized as Z-scores (Fig. 2A).Identification of mutant strains with perturbed lipid phenotypes
Deletion mutants with perturbed lipid phenotypes were identified using robust principal component analysis. This analysis was performed separately for mutant strains classified as either in the exponential or the stationary phase. I% values were used for the analysis. Missing values were substituted with zero. Calculations were performed using R software using the PcaHubert function from the rrcov package for robust multivariate analysis.34,35 Variables were scaled to have unit variance using median absolute deviation function. The number of principal components was selected based on scree-plots and was set to 4 for strains in the stationary phase and to 3 for strains in the exponential phase. PcaHubert function calculated orthogonal distance scores for each strains and the orthogonal distance cut-off value which was used to define the hit strains with perturbed lipid phenotypes. These strains were subjected to second round screening.Second round screening: liquid culturing
Liquid culture experiments of candidate mutant strains with perturbed lipid phenotypes and reference strains were performed at 30 °C with synthetic complete medium containing 2% glucose and supplemented with 100 μM inositol and 100 μM choline. The yeast strains were precultured for 24 hours, diluted to 0.2 OD600 units per ml and cultured for another 24 hours until collection of samples in the stationary phase (4.0–4.5 OD600 units per ml). Cells were washed with 155 mM ammonium acetate and stored at −80 °C until lipid extraction.Lipid extraction at 4 °C of samples obtained by liquid culturing
Samples from liquid cultures were subjected to two-step lipid extraction as previously described.4 Briefly, yeast cells were resuspended in 1 ml of 155 mM ammonium acetate and disrupted using glass bead lysis. Aliquots of cell lysates were diluted to 0.4 OD600 units in 200 μl and mixed with 17 μl of internal lipid standard mixture containing cholesterol-D7, CE 19:0, TAG 17:1/17:1/17:1, DAG 19:0/19:0, LPA O-16:0, PA 17:0/14:1, LPS 17:1, PS 17:0/20:4, LPE O-16:0, PE O-20:0/O-20:0, LPC O-17:0, PC 18:3/18:3, LPI 17:1, PI 17:0/14:1, PG 17:0/14:1, CL 14:0/14:0/14:0/14:0, Cer 18:1;2/17:0;0, IPC 18:0;2/26:0;0, MIPC 18:0;2/26:0;0 and M(IP)2C 18:0;2/26:0;0. Samples were extracted with 990 μl of chloroform/methanol (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 V/V) for 2 hours. The lower organic 15
1 V/V) for 2 hours. The lower organic 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1-phase was collected and subjected to vacuum evaporation. The remaining aqueous phase was re-extracted with 990 μl of chloroform/methanol (2
1-phase was collected and subjected to vacuum evaporation. The remaining aqueous phase was re-extracted with 990 μl of chloroform/methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 V/V) for 1 h. The lower organic 2
1 V/V) for 1 h. The lower organic 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1-phase was collected and subjected to vacuum evaporation.
1-phase was collected and subjected to vacuum evaporation.
Mass spectrometric lipid analysis and data processing for samples obtained by liquid culturing
The 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1- and 2
1- and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1-phase lipid extracts were dissolved in 100 μl of chloroform/methanol (1
1-phase lipid extracts were dissolved in 100 μl of chloroform/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) and analysed by direct infusion on a LTQ Orbitrap XL instrument equipped with the robotic TriVersa NanoMate ion source as previously described.4,29 Sterols were analyzed after chemical sulfation of the 15
2, v/v) and analysed by direct infusion on a LTQ Orbitrap XL instrument equipped with the robotic TriVersa NanoMate ion source as previously described.4,29 Sterols were analyzed after chemical sulfation of the 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 phase extract.24 The molar amount of lipid species were determined using the spiked-in internal standards and converted to mol% as previously described.4,29 For each strain, two technical replicates of a single lipid extract were analysed.
1 phase extract.24 The molar amount of lipid species were determined using the spiked-in internal standards and converted to mol% as previously described.4,29 For each strain, two technical replicates of a single lipid extract were analysed.
SoamD calculation
A SoamD (sum of absolute mol% difference) score was calculated for each deletion mutant subjected to second round screening. SoamD was calculated as:where N is the number of lipids, mol%i,j is the mol% value of lipid j in deletion mutant i and mol%BY4742,j is the mol% value of lipid j in the reference strain BY4742. The score is only applicable to experiments performed with spike-in of internal standards.
Results and discussion
A high-throughput platform for lipid phenotyping
To establish a resource for identifying proteins with potential function in lipid homeostasis we developed a high-throughput lipidomics platform for quantitative high-content screening of yeast mutant libraries. First, we compiled a deletion library in 96-well format covering 178 strains divided into three groups: (group A) 168 deletion mutants of genes encoding transcriptional regulators; (group B) 8 deletion mutants of genes encoding proteins with unknown function and predicted to be involved in lipid homeostasis; and (group C) control strains including the BY4742 reference and the elo2Δ mutant with defective fatty acid elongase activity36 (Table S1, ESI†). The group A strains were shortlisted based on GO annotations related to transcriptional activity. The group B strains were selected from an interaction network enriched with proteins involved in lipid metabolism and function (Fig. S1, ESI†). This network was compiled using the GeneMANIA function prediction algorithm31 that queried available protein–protein interaction data in the BioGRID database32 using a query list of lipid-related genes (Table S2, ESI†). Selected candidate proteins were all annotated as ‘biological process unknown’ in SGD30 (Table S3, ESI†).A key feature of high-throughput lipidomic screening is the ability to identify mutants with pronounced differences in lipid composition compared to reference strains or the average of the library. Differences in lipid composition are most accurately determined by absolute quantification of lipid species where the intensities of detected endogenous lipid species are normalized to the intensities and amounts of appropriate internal standards.4,23,29,37,38 The concentration of lipid species can be expressed as the molar abundance of lipid species relative to all monitored lipid species (i.e. mol%). Notably, absolute quantification on a lipidome-wide level requires spiking samples with ∼25 synthetic internal lipid standards, some of which are expensive or require cumbersome approaches to purify. Moreover, the workflow requires a dedicated two-step lipid extraction procedure that is difficult to execute in 96-well plate format. Thus, executing a lipidomic screening across hundreds of yeast strains utilizing absolute quantification on a lipidome-wide level is a challenging undertaking. To combat these technical and economical drawbacks we designed the screening platform to include two rounds of screening. A first round screening was designed for rapid lipid profiling across all shortlisted strains in the deletion library while the second round of screening was designed for comprehensive lipidome quantification in deletion mutants with perturbed lipid phenotypes identified in the first round screening.
For the first round screening, we devised and validated a lipidomic proxy supported by the comprehensive lipidome coverage obtained by high-resolution Orbitrap mass analysis. Lipidomic profiling in positive ion mode allows sensitive analysis of PC, LPC, PE, LPE, DAG, TAG, SE and Cer species. In addition, negative ion mode analysis allows monitoring of PI, LPI, PA, LPA, PS, LPS and IPC species. As such, the acquisition of spectral data for the same sample in both positive and negative ion mode allows monitoring of lipid species abundance by intensity profiling using the proxy “Intensity%” (I%) calculated as the intensity of a given lipid species divided by the sum of all monitored lipid intensities monitored within a given ion mode (i.e. positive or negative).
To benchmark the proxy for lipidomic screening, we compared the abundance of lipid species monitored by I% and mol% (Fig. 1A). To this end, we performed a comprehensive lipidomic analysis of nine yeast strains using the workflow for absolute quantification of lipid species as applied for the second round screening and in general for comparative lipidomic analysis.4,29 The abundances of endogenous lipid species from the same strain were expressed as both mol% (using internal standard information) and I% as outlined above. Our analysis demonstrated a linear correlation between the absolute abundance of lipid species and the lipid levels monitored by I%. Importantly, we observed that lipid species with high absolute mol% values displayed high I% values, and vice versa, lipid species with low mol% values displayed low I% values. Based on this result, we concluded that I% is a valid proxy for assessing the abundance of lipid species and mapping differences in lipid composition by high-throughput lipidomic screening. We note that identified mutants with perturbed lipid composition based on I% should be further investigated using the second round screening approach as this uses the more accurate absolute quantification on a lipidome-wide level. In addition, the second round screening should be performed since the first round lipidomic screening approach does not support quantification of free sterols as these analytes require additional chemical derivatization for quantification.4,29
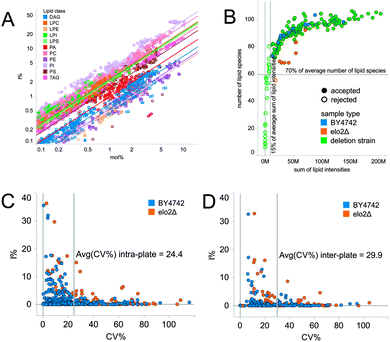 | ||
| Fig. 1 Validation of the shotgun lipidomic screening platform. (A) Correlation between I% and mol%. A lipidomics experiment of 9 yeast strains (used for the second round screening, see Fig. 4) was executed using appropriate internal standards to allow absolute quantification of lipid species (expressed as mol%). The same dataset was used for intensity profiling (expressed as I%). Lines correspond to linear correlation between mol% and I% values for endogenous lipid species of the same class. The average R2 value is 0.94. (B) Quality control plot with criteria for rejecting poor quality samples. Strains with less than 70% of the average number of detected lipid species and/or less than 15% of the average total intensity were excluded from subsequent analysis. Only negative mode data are shown. Positive mode data were filtered in the same way (data not shown). (C) Average intra-plate coefficient of variation. (D) Average inter-plate coefficient of variation. Inter-plate coefficient of variation reflects technical, within plate, plate-to-plate and biological variation of replicated measurements. | ||
Lipid profiling and data filtering for first round screening
All 178 strains in the shortlisted deletion library were subjected to the first round screening. Strains were cultured on synthetic complete solid medium in 96-well format for 24 hours in accordance with metabolomic studies.39,40 Cells were harvested, subjected to cell lysis and single-step lipid extraction in 96-well format followed by shotgun lipidomic analysis using a robotic nanoelectrospray device and a high-resolution Orbitrap mass spectrometer.4,28,41 The first round screening afforded comprehensive lipidomic analysis of 96 samples in approx. 12 hours. Detected lipid species were identified using ALEX software.27Next, we performed a quality control procedure to ensure the reliability of lipidome data across the 178 input strains. First, we introduced a filter to reject all strains featuring (i) less than 70% of the average number of detected lipid species and (ii) less than 15% of the average total lipid intensity (Fig. 1B). Using this approach, we passed 128 strains having on average 120 detected lipid species. We note that rejected strains were due to technical issues (e.g. poor ion spray) and poor growth.
To assess the precision of the lipidome data we evaluated the reproducibility of I% using biological replicates of the control strains BY4742 (n = 8) and elo2Δ (n = 7) distributed over three 96-well plates, and grown and analyzed together with shortlisted deletion mutants. The coefficient of variation (CV) for each lipid species detected in these control strains was determined (Fig. 1C and D). The average inter-plate CV of I% for lipid species was 30%. In comparison, the average intra-plate CV of I% was 24%. In addition, we observed a linear correlation between I% in biological replicates of the control strains (Pearson correlation R-square ≥ 0.973, p-value < 0.0001). We note that the CV values were determined for biological replicates of control strains grown for a fixed time rather than until the strains reached a definite cell amount. Hence, one can expect variation in the growth of individual strains which contributes to the relatively high CV values. We note that the applied culturing strategy is commonly used for large-scale metabolomics screening.39,40 Importantly, for the identification of mutant strains with altered lipid phenotypes we employed a multivariate method (described below) that differentiates mutant strains based on the composite of all lipid species I% values rather than the difference between I% values of single lipid species. Furthermore, the multivariate method differentiates lipid profiles across all surveyed strains instead of referencing only the control BY4742 strain. Using this approach minimizes the seemingly adverse impact of the relatively high CV values. Importantly, this approach successfully identified all replicates of the control mutant strain elo2Δ (i.e. no false negative identifications of elo2Δ) as having a perturbed lipid profile (Fig. 3A). We note that the lipid phenotype of elo2Δ is only modestly different from BY4742 as compared to elo3Δ.4 Moreover, the approach did not identify any of the replicates of the control BY4742 strain to display altered lipidome composition (i.e. no false positive identifications of BY4742). Based on these results we conclude that the first round lipidomic screening approach is a valid tool for surveying the lipid profile across hundreds of yeast strains.
Classification of deletion strains into growth phases
Yeast strains employ different lipid metabolic programs during exponential growth and the stationary phase, which can result in false-positive identification of lipid phenotypes.29 Notably, the yeast lipidome features high levels of glycerophospholipids during exponential growth which become offset by predominately TAG species and an increase in the chain length of fatty acid moieties in the stationary phase. In order to support accurate identification of mutants harbouring defects in lipid metabolism and not differences related to the growth phase, we executed the first round screening using 24 hours of culturing in order to allow ample time for all shortlisted deletion strains to enter the stationary phase. Moreover, we executed a parallel lipidomic analysis of exponential and stationary BY4742 cells cultured in liquid medium. Combining these two datasets and average linkage agglomerative hierarchical clustering allowed us to group lipid profiles of mutants from the deletion library into two clusters corresponding to cells in the exponential phase or the stationary phase based on lipid class composition and the species profile (Fig. 2A). As expected, majority of the deletion mutants (n = 116) displayed lipid profiles corresponding to the stationary phase having high levels of TAG species and glycerophospholipid species with longer chain fatty acid moieties as compared to exponential phase cells (Fig. 2B). In comparison, only a few strains (n = 12) displayed a lipidome composition similar to cells in the exponential phase. Having delineated the growth-dependent effects, we subsequently surveyed each group of strains separately for altered lipid metabolic phenotypes using multivariate analysis as outlined below.Identification of deletion strains with perturbed lipid phenotype
It has been demonstrated by recent genomics screening that identification of mutant strains with altered phenotypic traits can be efficiently achieved by referencing the whole collection of analyzed strains rather than comparison to a control strain.11–13 The advantage of such an approach is the possibility of using a higher number of strains for better estimation of technical and biological variation. Consequently, mutants with pronounced phenotypic traits can be identified more accurately. Here we applied a similar strategy for identification of deletion mutants with perturbed lipid phenotypes that is based on “robust principal component analysis”.42 Conventional principal component analysis can be an effective tool to identify key lipid features in multivariate lipidomic datasets.28 However, its efficacy can easily be hampered by outlying samples and lipid species. In contrast, the robust variant overcomes the limitation of sensitivity toward outliers by replacing the covariance matrix used for conventional principal component analysis with a robust covariance estimation.43 Consequently, the robust principal component analysis is better suited for identifying pronounced phenotypic alterations in deletion mutants rather than differences caused by technical and biological variation The robust principal component analysis reduces data dimensionality by computing principal components that explain a maximum amount of observed lipid phenotypic differences across all surveyed strains, and produces a diagnostic plot that classifies strains according to the magnitude and the similarities of lipid phenotypes (Fig. 3A). Strains that display a common pattern of changes but exhibit higher differences yield a higher score distance on the x-axis of the diagnostic plot. Strains that display uncommon differences that cannot be explained by the principal components receive high orthogonal distance scores displayed on the y-axis of the plot. Effectively, this approach allowed us to identify 11 mutant strains having higher orthogonal distances as compared to majority of the surveyed strains, and thus, potentially harbouring altered lipid phenotypes (Fig. 3A). The identified strains were all part of the stationary phase group (Fig. 2A) and represent ∼9% of all shortlisted strains in the deletion library. From the 11 identified candidates, 9 strains were deletion mutants of transcriptional regulators (cha4Δ, kar4Δ, met31Δ, mga2Δ, rrn10Δ, rsf2Δ, sir1Δ, sut2Δ, ume6Δ) and 2 were deletion mutants of genes encoding proteins predicted to play a role in lipid homeostasis (yjr015wΔ, ybr141cΔ). In addition, the robust principal component analysis identified all replicates of the control mutant elo2Δ. We note that replicates of BY4742 have relatively high score distances. This attribute can potentially be linked to the fact that the BY4742 strain is devoid of the kanamycin resistance cassette present in deletion strains and potentially able to affect cellular fitness44 or that BY4742 has not been subject to the same genetic selection as the mutant strains. Importantly, the orthogonal distance score for BY4742 replicates was not high and thereby illustrating no major lipidomic differences compared to majority of the deletion mutants.A prominent hit of the first round screening was the transcriptional regulator MGA2 (Fig. 3A). Mga2p is an endoplasmic reticulum membrane protein involved in the regulation of OLE1 transcription.10 Ole1p is the only fatty acid desaturase in S. cerevisiae and is therefore essential for the synthesis of monounsaturated fatty acids.45 Deletion of the MGA2 gene reduces the expression of Ole1p which results in lower levels of the monounsaturated fatty acids C16:1 and C18:1.46 Consequently, the mga2Δ strain synthesizes elevated amounts of lipids having fewer double bonds as compared to the control strain BY4742 (Fig. 3B). Notably, the first round screening showed that mga2Δ synthesizes primarily TAG species with a total of two double bonds and elevated levels of PC species having a single double bond (e.g. PC 32:1). Based on the ability to identify known constituents of the lipid metabolic network (i.e. mga2Δ and elo2Δ), we conclude that the first round lipidomic screening is a valid tool for identification of mutant strains with perturbed lipid phenotypic traits. We noted that deletion mutants for transcription factors involved in lipid metabolism Ino2p and Ino4p did not cause pronounced changes in lipid composition because the growth medium was supplemented with inositol and choline, which alleviate the phenotype of these deletions. The strain devoid of the transcriptional regulator Opi1p was excluded during the quality control procedure.
Second round screening of deletion strains with lipid phenotypes
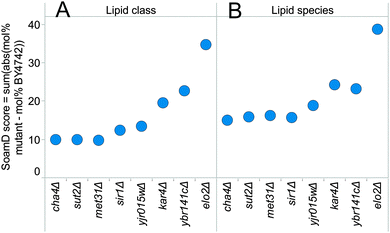
In order to further substantiate the lipid phenotypes of identified strains we executed a second round of lipidomic screening using the accurate and comprehensive workflow for absolute quantification of lipid species.4,29 To this end, we performed an extensive lipidome analysis of 7 deletion mutants, control strains elo2Δ and gup1Δ, and the reference strain BY4742 (Table S4, ESI†). The strains were cultured in synthetic complete liquid medium for 24 hours to allow cells to enter the stationary phase. Quantitative lipidomic analysis was performed using spike-in of internal standards and included the quantification of ergosterol and inositol-containing sphingolipids which were not monitored in the first round screening. To rank the lipid metabolic phenotypes we made use of a scoring algorithm that calculates the sum of absolute mol% difference relative to BY4742 (SoamD). This score was applied to rank the mutant strains according to the magnitude of the differences in lipid species and lipid class composition as compared to the reference strain BY4742 (Fig. 4). Using this approach we observed that the mutant elo2Δ harboured the most pronounced differences in global lipid composition followed by ybr141cΔ, kar4Δ and yjr015wΔ. The lipid phenotypes of ybr141cΔ, kar4Δ and yjr015wΔ will be discussed below.YJR015W has a plausible role in GPI-anchor synthesis
Yjr015wp is a protein of unknown function that localizes to the ER.47 It is predicted to have 6 transmembrane domains and function as a membrane transporter.48 The interaction landscape of YJR015W shows a direct interaction with key enzymes in fatty acid elongation and sphingolipid metabolism IFA38 and SUR4, and two enzymes involved in glycosylphosphatidylinositol (GPI) anchor synthesis GPI16 and GUP1 (Fig. 5A). All four interactors are transmembrane proteins that localize to the ER.16,49 Ifa38p and Sur4p are components of the elongase complex that synthesizes C26:0 fatty acid for Cer synthesis and remodeling of GPI-anchors via the O-acyl-transferase Gup1p. Gpi16p is a subunit of the transamidase complex that adds GPI-anchors to newly synthesized proteins. Notably, GPI-anchored proteins in S. cerevisiae comprise either a glycerophospholipid PI species or a sphingolipid IPC species. In addition, YJR015W also interacts with enzymes responsible for N-linked glycosylation and machinery involved in ER to Golgi vesicle transport (Fig. 5A). Taken together, these interactions support the notion that Yjr015wp is potentially involved in GPI-anchor synthesis. Based on this prediction we included gup1Δ as a control strain in the comparative lipidomic analysis.The lipid phenotype of yjr015wΔ revealed a distinct set of perturbed sphingolipid features (Fig. 5B and C). The yjr015wΔ lipidome showed increased levels of 46:0;4 sphingolipid species being offset by a reduction in 44:0;5 sphingolipid species. The 46:0;4 species correspond to a sphingolipid composed of a C20 phytosphingosine and an amide-linked hydroxylated C26:0 fatty acid moiety.4,50 In comparison, the 44:0;5 species correspond to sphingolipid having a C18 phytosphingosine and a C26:0 fatty acid moiety with two hydroxyl groups. The hydroxylase that inserts the second hydroxyl group into the C26 fatty acid moiety as well as its position in the fatty acid chain and the molecular function of the produced sphingolipid molecule are unknown.51,52 It is unlikely that the sphingolipid phenotype of yjr015wΔ is attributed to a reduced activity of the fatty acid elongation complex because inactivation of the interactor Sur4p would shorten the fatty acid chain length of sphingolipids.4,36 Instead, the perturbation of sphingolipid hydroxylation profile could be due to reduced activity of the unknown hydroxylase or selective utilization of IPC 44:0;5 species for remodeling of GPI-anchors. Interestingly, inactivation of GPI-anchor remodeling in the gup1Δ mutants coincides with a reduction in IPC 44:0;5 (termed IPC-D in thin-layer chromatographic analysis) and increased incorporation of a base resistant anchor lipid with chromatographic properties similar to IPC 44:0;5.53 In addition, it has been observed that GPI-anchor proteins in gup1Δ cells comprised lower levels of IPC 44:0;4. Taken together, these data support the observed reduction in all sphingolipid 44:0;5 species and elevated levels of 46:0;4 species in the yjr015wΔ lipidome (Fig. 5B). Our analysis showed that the yjr015wΔ lipidome partially phenocopied the gup1Δ lipidome with respect to the top 10 decreasing lipid species (Fig. 5B and D). In both mutant strains, the 44:0;5 sphingolipid species were among the most reduced lipid species. In contrast, the observed top 10 increased lipid species were only partially conserved in the two mutant strains. As for the lipid class phenotype we also observed a strong similarity between the yjr015wΔ and gup1Δ lipidome. Based on the distinct lipid phenotype of yjr015wΔ, its similarity to the gup1Δ lipidome and the interaction landscape of YJR015W we propose that Yjr015wp might play a functional role in modulation of GPI-anchor synthesis. We note that in order to reveal the exact role of YJR015W an additional study of the yjr015wΔ strain and the Yjr015w protein is required.
Ybr141cp – a putative methyltransferase involved in sterol lipid metabolism
YBR141C encodes a putative methyltransferase that localizes to the nucleolus.47 A recent bioinformatic study of yeast methyltransferases predicted Ybr141cp to contain a Rossmann-like catalytic domain similar to the sterol methyltransferase Erg6p that converts zymosterol to fecosterol in the ergosterol biosynthetic pathway.54 The catalytic Rossmann-like domain spans methyltransferases with diverse substrate specificities including sterols, proteins, RNA and other small molecules. As such, Ybr141cp was predicted to use rRNA or tRNA as the substrate,54 albeit this has not been experimentally verified. In addition, a number of proteins devoid of methyltransferase activity and featuring the Rossmann-like domain have been identified. These proteins include the transcription factor Kar4p and the mitochondrial RNA polymerase specificity factor Mtf1p.54,55 The interaction network of YBR141C shows a link to lipid metabolism via a physical interaction with Vps74p (Fig. S1, ESI†), a phosphoinositide-binding protein involved in localizing glycosyltransferases in the Golgi.56The lipid phenotype of ybr141cΔ showed a pronounced increase of ergosterol esters offset by a reduction of ergosterol (Fig. 6A and B). In addition, the ybr141cΔ lipidome showed a concomitant increase in TAG levels and a reduction in all membrane glycerophospholipids. The reason for this distinct lipid phenotype is at the present time unclear given the limited information about YBR141C function. Interestingly, a similar perturbation of sterol esters and free sterol levels was observed when inactivating ERG6.57 Moreover, chemical genomic data show that YBR141C has a co-fitness interaction with the major sterol acyl-transferase ARE2.58 Although the information about YBR141C function is limited, our results provide a framework for testing the functional role of Ybr141cp in sterol metabolism.
Kar4p – a transcription factor linked to nuclear membrane dynamics
KAR4 encodes a transcription factor required for nuclear fusion during yeast mating and possibly other functions during vegetative growth.59–62 Kar4p exists as two isoforms; a constitutive 38.5 kD protein (Kar4p-long) that predominates during vegetative growth and a 35.5 kD protein (Kar4p-short) that is induced during mating.61 During the mating process, Kar4p-short acts together with the transcription factor Ste12p to induce the expression of KAR3 and CIK1 that encode a motor protein complex required for congression of nuclei prior to nuclear membrane fusion.60,62 During vegetative growth, Kar4p-long expression is up-regulated in the G1 phase of the cell cycle and implicated in constitutive expression of more than 50 genes.60,62 The kar4Δ deletion mutant displays a slow growth phenotype attributed to a short G1 pause during vegetative growth, and a pronounced defect in nuclear congression during mating that phenocopies the absence of KAR3 and CIK1.59Given the functional role of Kar4p in nuclear fusion, it is plausible that Kar4p is also involved in regulating nuclear membrane dynamics during the cell cycle. Notably, perturbing lipid metabolism has previously been shown to compromise nuclear membrane growth and function.63 Deletion of the PA phosphatase Pah1p and components of its regulatory complex Nem1p-Spo7p reduces PA to DAG conversion and causes nuclear membrane expansion.64,65 In addition, overexpression of the nuclear/ER-localized DAG kinase Dgk1p phenocopies the Pah1p deficiency66 indicating that regulation of the composition of DAG, PA and other glycerophospholipids is important for nuclear membrane dynamics. Conversely, deletion of integral nuclear membrane-ER proteins Brr6 and Apq12 precipitate defects in nuclear pore complex assembly, sterol metabolism and lipid droplet morphology.67
The lipid phenotype of kar4Δ showed a pronounced increase in DAG and SE species being offset by a reduction in primarily PE species and ergosterol (Fig. 7). Interestingly, this lipid phenotype is reminiscent of the lipid compositions associated with nuclear membrane defects observed in the previous studies. The elevated DAG levels and reduction in PE levels are similar to the effects of overexpressing Pah1p,68 whereas the increased levels of SE and reduced levels of ergosterol resemble effects of defective nuclear membrane growth in the pah1Δ deletion mutant.69 This apparent combination of perturbed lipid features indicates that inactivation of Kar4p potentially fails to prompt inhibition of Pah1p activity, which in turn channels PA into DAG production instead of synthesis of PE and other glycerophospholipids for membrane expansion. The accumulation of SE could be a secondary effect of kar4Δ cells trying to synchronize the rate of ergosterol biosynthesis and secretory vesicle flow under the reduced vegetative growth rate. We here note that the exact function of Kar4p action during vegetative growth requires further characterization of the kar4Δ strain and Kar4 protein.
Conclusion
Existing high throughput screening methodologies for identification of proteins with a function in lipid metabolism provide only limited phenotypic information. Epistatic miniarray profiling and protein–protein interaction assays are based on monitoring growth fitness (i.e. colony size) which allows mapping functionally related genes and proteins, respectively.11–13 Alternatively, microscopy-based screening can be used for identifying mutant strains with perturbed lipid droplet dynamics based on altered lipid droplet size and morphology.70,71 In contrast, detailed assessment of cellular lipid composition demands dedicated methods such as mass spectrometry-based lipidomics. Several groups have recently reported workflows for lipidomics analysis in yeast, but their application has so far been limited to characterisation of only a few mutant strains or a reference strain grown under different conditions.4,29,72,73In the current study, we designed a platform for high-content lipidomic screening of yeast mutant libraries. We combined culturing and lipid extraction in 96-well format, automated direct infusion nanoelectrospray ionization, high-resolution Orbitrap mass spectrometry and a novel data processing framework to support lipid phenotyping across hundreds of S. cerevisiae mutants. The screening platform was designed to include two rounds of screening. A first round screening was executed for rapid lipid profiling across all shortlisted strains in the deletion library while a second round of screening was conducted for more comprehensive lipidome quantification of deletion mutants with perturbed lipid phenotypes identified in the first round screening. To our knowledge, this is the first assessment of lipidomic phenotypes across hundreds of mutants in a single screen. Notably, our platform extends the palette of analytical techniques available for functional genomics studies aimed at uncovering proteins with previously unknown function in lipid metabolism and regulators of global lipid homeostasis. The technology affords a multidimensional survey of physiological lipid parameters that helps explore uncharacterized proteins and propose valuable hypothesis for mechanistic biochemical follow-up experiments. Our case study of a library covering deletion mutants of genes with predicted function in lipid metabolism and transcriptional regulators revealed three poorly characterized genes that precipitate distinct lipid metabolic phenotypes upon deletion. In conclusion, the high-throughput lipidomic screening platform described herein is a valid and complementary tool for high-content analysis of yeast mutant libraries.
Acknowledgements
We are grateful to Nils J. Færgeman, Robin W. Klemm, Ole N. Jensen and Jens Knudsen for productive discussions and access to equipment. We thank Valentin Todorov for valuable advice on multivariate methods. We thank the reviewers for their constructive comments on the manuscript. This work was supported by the Danish Council for Independent Research | Natural Sciences (09-072484; 10-094213) and Lundbeckfonden (R44-A4342).Notes and references
- A. Shevchenko and K. Simons, Nat. Rev. Mol. Cell Biol., 2010, 11, 593–598 CrossRef CAS PubMed.
- G. van Meer, EMBO J., 2005, 24, 3159–3165 CrossRef CAS PubMed.
- E. Fahy, S. Subramaniam, R. C. Murphy, M. Nishijima, C. R. Raetz, T. Shimizu, F. Spener, G. van Meer, M. J. Wakelam and E. A. Dennis, J. Lipid Res., 2009, 50(suppl), S9–S14 Search PubMed.
- C. S. Ejsing, J. L. Sampaio, V. Surendranath, E. Duchoslav, K. Ekroos, R. W. Klemm, K. Simons and A. Shevchenko, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2136–2141 CrossRef CAS PubMed.
- M. P. Wymann and R. Schneiter, Nat. Rev. Mol. Cell Biol., 2008, 9, 162–176 CrossRef CAS PubMed.
- R. C. Dickson, J. Lipid Res., 2008, 49, 909–921 CrossRef CAS PubMed.
- G. M. Carman and G. S. Han, Annu. Rev. Biochem., 2011, 80, 859–883 CrossRef CAS PubMed.
- M. Kanehisa and S. Goto, Nucleic Acids Res., 2000, 28, 27–30 CrossRef CAS PubMed.
- S. A. Henry, S. D. Kohlwein and G. M. Carman, Genetics, 2012, 190, 317–349 CrossRef CAS PubMed.
- R. Chellappa, P. Kandasamy, C. S. Oh, Y. Jiang, M. Vemula and C. E. Martin, J. Biol. Chem., 2001, 276, 43548–43556 CrossRef CAS PubMed.
- M. Costanzo, A. Baryshnikova, J. Bellay, Y. Kim, E. D. Spear, C. S. Sevier, H. Ding, J. L. Koh, K. Toufighi, S. Mostafavi, J. Prinz, R. P. St Onge, B. VanderSluis, T. Makhnevych, F. J. Vizeacoumar, S. Alizadeh, S. Bahr, R. L. Brost, Y. Chen, M. Cokol, R. Deshpande, Z. Li, Z. Y. Lin, W. Liang, M. Marback, J. Paw, B. J. San Luis, E. Shuteriqi, A. H. Tong, N. van Dyk, I. M. Wallace, J. A. Whitney, M. T. Weirauch, G. Zhong, H. Zhu, W. A. Houry, M. Brudno, S. Ragibizadeh, B. Papp, C. Pal, F. P. Roth, G. Giaever, C. Nislow, O. G. Troyanskaya, H. Bussey, G. D. Bader, A. C. Gingras, Q. D. Morris, P. M. Kim, C. A. Kaiser, C. L. Myers, B. J. Andrews and C. Boone, Science, 2010, 327, 425–431 CrossRef CAS PubMed.
- M. Schuldiner, S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, J. S. Weissman and N. J. Krogan, Cell, 2005, 123, 507–519 CrossRef CAS PubMed.
- K. Tarassov, V. Messier, C. R. Landry, S. Radinovic, M. M. Serna Molina, I. Shames, Y. Malitskaya, J. Vogel, H. Bussey and S. W. Michnick, Science, 2008, 320, 1465–1470 CrossRef CAS PubMed.
- J. P. Miller, R. S. Lo, A. Ben-Hur, C. Desmarais, I. Stagljar, W. S. Noble and S. Fields, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12123–12128 CrossRef CAS PubMed.
- S. R. Collins, P. Kemmeren, X. C. Zhao, J. F. Greenblatt, F. Spencer, F. C. Holstege, J. S. Weissman and N. J. Krogan, Mol. Cell. Proteomics, 2007, 6, 439–450 CAS.
- V. Denic and J. S. Weissman, Cell, 2007, 130, 663–677 CrossRef CAS PubMed.
- D. K. Breslow, S. R. Collins, B. Bodenmiller, R. Aebersold, K. Simons, A. Shevchenko, C. S. Ejsing and J. S. Weissman, Nature, 2010, 463, 1048–1053 CrossRef CAS PubMed.
- P. S. Aguilar, F. Frohlich, M. Rehman, M. Shales, I. Ulitsky, A. Olivera-Couto, H. Braberg, R. Shamir, P. Walter, M. Mann, C. S. Ejsing, N. J. Krogan and T. C. Walther, Nat. Struct. Mol. Biol., 2010, 17, 901–908 CAS.
- P. I. Wang and E. M. Marcotte, J. Proteomics, 2010, 73, 2277–2289 CrossRef CAS PubMed.
- D. C. Hess, C. L. Myers, C. Huttenhower, M. A. Hibbs, A. P. Hayes, J. Paw, J. J. Clore, R. M. Mendoza, B. S. Luis, C. Nislow, G. Giaever, M. Costanzo, O. G. Troyanskaya and A. A. Caudy, PLoS Genet., 2009, 5, e1000407 Search PubMed.
- M. D. Chikina, C. Huttenhower, C. T. Murphy and O. G. Troyanskaya, PLoS Comput. Biol., 2009, 5, e1000417 Search PubMed.
- B. Brugger, B. Glass, P. Haberkant, I. Leibrecht, F. T. Wieland and H. G. Krausslich, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2641–2646 CrossRef PubMed.
- R. W. Klemm, C. S. Ejsing, M. A. Surma, H. J. Kaiser, M. J. Gerl, J. L. Sampaio, Q. de Robillard, C. Ferguson, T. J. Proszynski, A. Shevchenko and K. Simons, J. Cell Biol., 2009, 185, 601–612 CAS.
- M. Carvalho, J. L. Sampaio, W. Palm, M. Brankatschk, S. Eaton and A. Shevchenko, Mol. Syst. Biol., 2012, 8, 600 CrossRef PubMed.
- C. Scholz, D. Parcej, C. S. Ejsing, H. Robenek, I. L. Urbatsch and R. Tampe, J. Biol. Chem., 2011, 286, 13346–13356 CrossRef PubMed.
- X. Han, K. Yang and R. W. Gross, Mass Spectrom. Rev., 2012, 31, 134–178 CrossRef CAS PubMed.
- P. Husen, K. Tarasov, M. Katafiasz, E. Sokol, J. Vogt, J. Baumgart, R. Nitsch, K. Ekroos and C. S. Ejsing, PLoS One, 2013, 8, e79736 CAS.
- D. Schwudke, J. T. Hannich, V. Surendranath, V. Grimard, T. Moehring, L. Burton, T. Kurzchalia and A. Shevchenko, Anal. Chem., 2007, 79, 4083–4093 CrossRef CAS PubMed.
- C. Klose, M. A. Surma, M. J. Gerl, F. Meyenhofer, A. Shevchenko and K. Simons, PLoS One, 2012, 7, e35063 CAS.
- J. M. Cherry, E. L. Hong, C. Amundsen, R. Balakrishnan, G. Binkley, E. T. Chan, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. R. Engel, D. G. Fisk, J. E. Hirschman, B. C. Hitz, K. Karra, C. J. Krieger, S. R. Miyasato, R. S. Nash, J. Park, M. S. Skrzypek, M. Simison, S. Weng and E. D. Wong, Nucleic Acids Res., 2012, 40, D700–D705 CrossRef CAS PubMed.
- D. Warde-Farley, S. L. Donaldson, O. Comes, K. Zuberi, R. Badrawi, P. Chao, M. Franz, C. Grouios, F. Kazi, C. T. Lopes, A. Maitland, S. Mostafavi, J. Montojo, Q. Shao, G. Wright, G. D. Bader and Q. Morris, Nucleic Acids Res., 2010, 38, W214–W220 CrossRef CAS PubMed.
- C. Stark, B. J. Breitkreutz, T. Reguly, L. Boucher, A. Breitkreutz and M. Tyers, Nucleic Acids Res., 2006, 34, D535–D539 CrossRef CAS PubMed.
- M. J. de Hoon, S. Imoto, J. Nolan and S. Miyano, Bioinformatics, 2004, 20, 1453–1454 CrossRef CAS PubMed.
- R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria, 2011.
- V. Todorov and P. Filzmoser, J. Stat. Software, 2009, 32, 1–47 Search PubMed.
- C. S. Oh, D. A. Toke, S. Mandala and C. E. Martin, J. Biol. Chem., 1997, 272, 17376–17384 CrossRef CAS PubMed.
- J. L. Sampaio, M. J. Gerl, C. Klose, C. S. Ejsing, H. Beug, K. Simons and A. Shevchenko, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1903–1907 CrossRef CAS PubMed.
- M. A. Surma, C. Klose, R. W. Klemm, C. S. Ejsing and K. Simons, Traffic, 2011, 12, 1139–1147 CrossRef CAS PubMed.
- S. J. Cooper, G. L. Finney, S. L. Brown, S. K. Nelson, J. Hesselberth, M. J. MacCoss and S. Fields, Genome Res., 2010, 20, 1288–1296 CrossRef CAS PubMed.
- J. Allen, H. M. Davey, D. Broadhurst, J. K. Heald, J. J. Rowland, S. G. Oliver and D. B. Kell, Nat. Biotechnol., 2003, 21, 692–696 CrossRef CAS PubMed.
- J. Folch, M. Lees and G. H. Sloane Stanley, J. Biol. Chem., 1957, 226, 497–509 CAS.
- M. Hubert and S. Engelen, Bioinformatics, 2004, 20, 1728–1736 CrossRef CAS PubMed.
- M. Hubert, P. J. Rousseeuw and K. Vanden Branden, Technometrics, 2005, 47, 64–79 CrossRef.
- R. Babazadeh, S. M. Jafari, M. Zackrisson, A. Blomberg, S. Hohmann, J. Warringer and M. Krantz, FEBS Lett., 2011, 585, 3907–3913 CrossRef CAS PubMed.
- J. E. Stukey, V. M. McDonough and C. E. Martin, J. Biol. Chem., 1989, 264, 16537–16544 CAS.
- M. A. Surma, C. Klose, D. Peng, M. Shales, C. Mrejen, A. Stefanko, H. Braberg, D. E. Gordon, D. Vorkel, C. S. Ejsing, R. Farese, Jr., K. Simons, N. J. Krogan and R. Ernst, Mol. Cell, 2013, 51, 519–530 CrossRef CAS PubMed.
- W. K. Huh, J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman and E. K. O'Shea, Nature, 2003, 425, 686–691 CrossRef CAS PubMed.
- B. De Hertogh, E. Carvajal, E. Talla, B. Dujon, P. Baret and A. Goffeau, Funct. Integr. Genomics, 2002, 2, 154–170 CrossRef CAS PubMed.
- P. Orlean and A. K. Menon, J. Lipid Res., 2007, 48, 993–1011 CrossRef CAS PubMed.
- C. S. Ejsing, T. Moehring, U. Bahr, E. Duchoslav, M. Karas, K. Simons and A. Shevchenko, J. Mass Spectrom., 2006, 41, 372–389 CrossRef CAS PubMed.
- R. Schneiter, BioEssays, 1999, 21, 1004–1010 CrossRef CAS PubMed.
- T. J. Beeler, D. Fu, J. Rivera, E. Monaghan, K. Gable and T. M. Dunn, MGG, Mol. Gen. Genet., 1997, 255, 570–579 CrossRef CAS PubMed.
- R. Bosson, M. Jaquenoud and A. Conzelmann, Mol. Biol. Cell, 2006, 17, 2636–2645 CrossRef CAS PubMed.
- T. Wlodarski, J. Kutner, J. Towpik, L. Knizewski, L. Rychlewski, A. Kudlicki, M. Rowicka, A. Dziembowski and K. Ginalski, PLoS One, 2011, 6, e23168 CAS.
- J. M. Bujnicki, M. Feder, M. Radlinska and R. M. Blumenthal, J. Mol. Evol., 2002, 55, 431–444 CrossRef CAS PubMed.
- P. Uetz, L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields and J. M. Rothberg, Nature, 2000, 403, 623–627 CrossRef CAS PubMed.
- M. T. McCammon, M. A. Hartmann, C. D. Bottema and L. W. Parks, J. Bacteriol., 1984, 157, 475–483 CAS.
- M. E. Hillenmeyer, E. Ericson, R. W. Davis, C. Nislow, D. Koller and G. Giaever, Genome Biol., 2010, 11, R30 CrossRef PubMed.
- L. J. Kurihara, C. T. Beh, M. Latterich, R. Schekman and M. D. Rose, J. Cell Biol., 1994, 126, 911–923 CrossRef CAS PubMed.
- L. J. Kurihara, B. G. Stewart, A. E. Gammie and M. D. Rose, Mol. Cell. Biol., 1996, 16, 3990–4002 CrossRef CAS PubMed.
- A. E. Gammie, B. G. Stewart, C. F. Scott and M. D. Rose, Mol. Cell. Biol., 1999, 19, 817–825 CrossRef CAS PubMed.
- R. Lahav, A. Gammie, S. Tavazoie and M. D. Rose, Mol. Cell. Biol., 2007, 27, 818–829 CrossRef CAS PubMed.
- S. Siniossoglou, Biochim. Biophys. Acta, 2013, 1831, 575–581 CrossRef CAS PubMed.
- S. Siniossoglou, H. Santos-Rosa, J. Rappsilber, M. Mann and E. Hurt, EMBO J., 1998, 17, 6449–6464 CrossRef CAS PubMed.
- H. Santos-Rosa, J. Leung, N. Grimsey, S. Peak-Chew and S. Siniossoglou, EMBO J., 2005, 24, 1931–1941 CrossRef CAS PubMed.
- G. S. Han, L. O'Hara, G. M. Carman and S. Siniossoglou, J. Biol. Chem., 2008, 283, 20433–20442 CrossRef CAS PubMed.
- C. A. Hodge, V. Choudhary, M. J. Wolyniak, J. J. Scarcelli, R. Schneiter and C. N. Cole, J. Cell Sci., 2010, 123, 141–151 CrossRef CAS PubMed.
- G. S. Han, S. Siniossoglou and G. M. Carman, J. Biol. Chem., 2007, 282, 37026–37035 CrossRef CAS PubMed.
- G. S. Han, W. I. Wu and G. M. Carman, J. Biol. Chem., 2006, 281, 9210–9218 CrossRef CAS PubMed.
- W. Fei, G. Shui, B. Gaeta, X. Du, L. Kuerschner, P. Li, A. J. Brown, M. R. Wenk, R. G. Parton and H. Yang, J. Cell Biol., 2008, 180, 473–482 CrossRef CAS PubMed.
- W. Fei, G. Shui, Y. Zhang, N. Krahmer, C. Ferguson, T. S. Kapterian, R. C. Lin, I. W. Dawes, A. J. Brown, P. Li, X. Huang, R. G. Parton, M. R. Wenk, T. C. Walther and H. Yang, PLoS Genet., 2011, 7, e1002201 CAS.
- P. Chumnanpuen, J. Zhang, I. Nookaew and J. Nielsen, Mol. Genet. Genomics, 2012, 287, 541–554 CrossRef CAS PubMed.
- G. Shui, X. L. Guan, C. P. Low, G. H. Chua, J. S. Goh, H. Yang and M. R. Wenk, Mol. BioSyst., 2010, 6, 1008–1017 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary Fig. S1. Supplementary Tables S1–4. See DOI: 10.1039/c3mb70599d |
| This journal is © The Royal Society of Chemistry 2014 |