Open Access Article
Open Access ArticleIntegrated lab-on-chip biosensing systems based on magnetic particle actuation – a comprehensive review
Alexander
van Reenen
ac,
Arthur M.
de Jong
ac,
Jaap M. J.
den Toonder
bc and
Menno W. J.
Prins
*acd
aDepartment of Applied Physics, Eindhoven University of Technology, Eindhoven, The Netherlands. E-mail: m.w.j.prins@tue.nl; menno.prins@philips.com
bDepartment of Mechanical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands
cInstitute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands
dPhilips Research, Royal Philips, Eindhoven, The Netherlands
First published on 10th March 2014
Abstract
The demand for easy to use and cost effective medical technologies inspires scientists to develop innovative lab-on-chip technologies for point-of-care in vitro diagnostic testing. To fulfill medical needs, the tests should be rapid, sensitive, quantitative, and miniaturizable, and need to integrate all steps from sample-in to result-out. Here, we review the use of magnetic particles actuated by magnetic fields to perform the different process steps that are required for integrated lab-on-chip diagnostic assays. We discuss the use of magnetic particles to mix fluids, to capture specific analytes, to concentrate analytes, to transfer analytes from one solution to another, to label analytes, to perform stringency and washing steps, and to probe biophysical properties of the analytes, distinguishing methodologies with fluid flow and without fluid flow (stationary microfluidics). Our review focuses on efforts to combine and integrate different magnetically actuated assay steps, with the vision that it will become possible in the future to realize integrated lab-on-chip biosensing assays in which all assay process steps are controlled and optimized by magnetic forces.
1 Introduction
The ageing population and its accompanying increase in chronic disease prevalence put high pressure on the healthcare system, which drives the need for easy-to-use and cost-effective medical technologies.1In vitro diagnostics (IVD) plays a large role in delivering healthcare: it makes up a few percent of a hospital's budget but leverages the majority of all critical decision-making such as admittance, discharge, and medication.2,3 Decentralizing diagnostic testing, i.e. point-of-care testing (POCT), is a growing segment in the IVD market. POCT reduces the turn-around times to the physician, resulting in faster treatment decisions, giving improved workflows and improving the quality of care.4 Furthermore, POCT devices create opportunities to perform testing in less expensive settings such as the doctor's office and the home. This creates the possibility to deliver cost-effective care, e.g. remotely monitoring the progress of patients, personalization of treatment, and reducing the number of visits needed to the hospital.Applications for which POCT is very relevant are, for example, the detection of protein markers to diagnose cardiac diseases and the detection of nucleic acid markers in case of infectious diseases. The detection of these biomarkers requires that POCT devices not only contain a sensing technology but can also perform all of the sample pretreatment steps that are required in the assay, thus becoming so-called lab-on-a-chip or micro-total-analysis systems,5 in which microfluidics plays an important role.6 As biomarkers are typically present at very low concentrations within complex samples that contain high concentrations of background material, the methodologies should be highly selective and accurate. In the case of protein biomarker detection, molecular selectivity can be obtained by making use of antibodies in immunoassays,7 and a high sensing accuracy can be achieved by introducing labels in the assay. In nucleic-acid assays, purification and biochemical amplification steps are typically applied.8
Concerning protein biomarker detection, several immunoassay sensing technologies have been developed, such as nanoparticle labeling,9–11 label-free electrical detection,12 fluorescence detection13,14 and oligonucleotide labeling combined with biochemical amplification.15 While the detection sensitivities of these technologies can be high, the integration of these platforms in cost-effective lab-on-chip devices is complicated because several active fluidic steps are required to enable sample-pretreatment,9–15 (bio)chemical development9,15 or washing using buffer fluids.9–15 Therefore, it is important to face the challenge of total integration16 and design solutions that facilitate all assay steps, from sample preparation to final detection.
For several decades, magnetic particles have been applied in pipette-based assays, ranging from manual assays for basic research to assays in high-throughput instruments for centralized laboratories.18 The main advantages of using magnetic particles† are that they have a large surface-to-volume ratio, they are conveniently biofunctionalized, and they can be manipulated by magnetic fields, thereby simplifying extraction and buffer replacement steps. Particles are commercially available with different sizes, magnetic properties and surface coatings. Most particles are synthesized by coprecipitation or thermal decomposition.19 The magnetic core can be composed of e.g. iron oxides (like magnetite Fe3O4 or maghemite γ-Fe2O3) or pure metals (like Fe and Co) or alloys (like FePt). The magnetic material is protected by a coating that can be polymeric or inorganic and that also serves as a starting point for bio-functionalization. When many separate magnetic grains are embedded inside a non-magnetic matrix,20 the particle as a whole can exhibit superparamagnetic behavior,21i.e. the particles are paramagnetic with a very high magnetic susceptibility.
The availability of magnetic particles and corresponding assay reagents has formed a solid starting point for explorations toward miniaturization, i.e. efforts to realize integrated and miniaturized technologies based on magnetic particles and small fluid volumes.19,22 On small scales, it is difficult in principle to manipulate fluids due to high flow resistances, dominance of capillary forces, and difficulties to achieve mixing at low Reynolds numbers. Yet, the manipulation of magnetic particles by magnetic fields scales favorably with system miniaturization, because close to the field generators the magnetic fields are strong, because magnetic field gradients are large in the vicinity of structures with high curvature, and in addition only short distances need to be travelled in miniaturized devices. This has led to the concept of stationary microfluidics,23–25 in which overall fluid manipulation is minimized and the control of assay steps is mainly effectuated by magnetic particles and magnetic forces.
Magnetic particles are highly versatile and have been studied for many process steps that are required for lab-on-chip diagnostic assays. Magnetic particles have been applied (see Fig. 1) to mix fluids, to selectively capture specific analytes (i.e. the biomarkers that need to be detected), to concentrate analytes, to transfer analytes from one solution to another, to label analytes, to perform stringency and washing steps, and to probe biophysical properties of the analytes. In this review, an overview of the accomplishments of magnetic particles in all of these functions will be given. The review is centered around the concept that the integration of point-of-care assays can be facilitated by using actuated magnetic particles. We describe key assay steps in which magnetic particle actuation can play a role: mixing (section 2), analyte capture (section 3), and analyte detection (section 4). These three process steps are essential in every assay based on molecular binding affinity. Thereafter, we summarize the status of the integration of the different magnetically actuated assay steps with the vision that in the future it will become possible to realize integrated lab-on-chip biosensing assays in which all assay processes are controlled and optimized by magnetic forces. We draw examples from studies reported in the peer-reviewed scientific literature including our own research papers. We focus on the application of magnetic actuation in immunoassays and somewhat less on nucleic-acid detection assays. Finally, we discuss the current challenges and possible directions for integrated biosensing based on actuated magnetic particles in microfluidic devices.
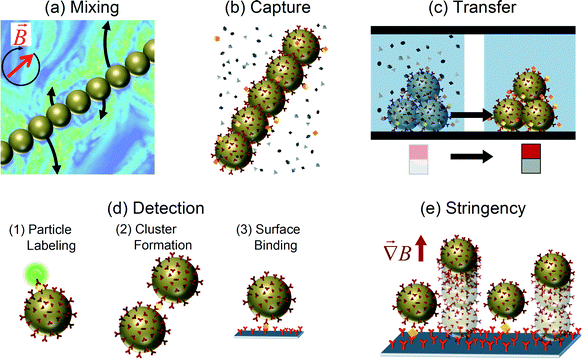 | ||
| Fig. 1 Application of magnetic particles in several process steps of a lab-on-chip diagnostic assay. Actuated by applied magnetic fields, magnetic particles have been used (a) to mix fluids, (b) to selectively capture specific analytes, (c) to transfer analytes to another fluid, (d1) to label particles for detection, (d2) to form clusters for detection, (d3) to induce surface binding for detection, and (e) to apply stringency forces in order to improve the signal-to-background ratio. (a) Adapted with permission from ref. 17 Copyright 2007 The American Physical Society. | ||
2 Microfluidic mixing using magnetic particles
The mixing of fluids has been a topic of long-standing interest in the microfluidics community. Due to the small dimensions of microfluidic devices, viscous forces are strong and result in slow and inefficient mixing. In most POCT devices, rapid mixing of two or more fluids or solutes is an essential step. Methods based on magnetic particles and magnetic fields have been investigated with the aim to improve microfluidic mixing. The methods can be separated in two classes: (i) mixing of fluid layers in a laminar fluid flow and (ii) mixing of fluid in a static fluid compartment, which will both be discussed in the following sections.2.1 Mixing a flowing fluid using magnetic particles
In the presence of a magnetic field, magnetic particles tend to form chains and other supra-particle structures due to the magnetic dipole–dipole interaction between the particles. Such magnetic structures can be used to stir fluids on the microscale. Hayes et al.26 found that applying a magnetic field to a suspension of superparamagnetic particles in a microfluidic channel causes the formation of dynamic and reversible self-assembled regularly-spaced supraparticle structures (see Fig. 2a). The formed structures or “plugs” could be rotated around all axes without losing their structural form. In addition, these plugs resisted deformation under pressure flow conditions and as such are able to influence fluid flow within a microchannel.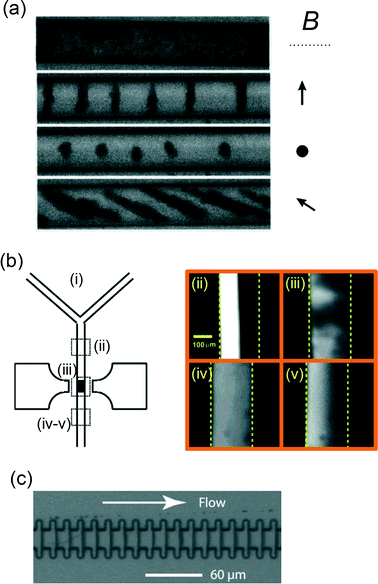 | ||
| Fig. 2 Examples of assembled magnetic particle structures to increase particle–fluid interactions within a microfluidic flow. (a) Formation of supraparticle structures of superparamagnetic particles (∅1–2 μm) within a microchannel (∅20 μm) for different field orientations, as indicated by the arrows. (b) Experimental evaluation of fluid mixing of parallel fluorescent and non-fluorescent streams within a channel by magnetically retained and actuated supra-particle structures. The fluorescence images (ii–v) on the right are taken at different locations as indicated in panel (i), i.e.: (ii) before mixing; (iii) during mixing; (iv) after mixing by a 20 Hz sinusoidal field; and (v) after mixing by a 5 Hz square-shaped field. (c) An optical image of a microchannel engineered to assemble and retain magnetic particles into 20 plugs across the channel. (a) Reprinted with permission from ref. 26 Copyright 2001 American Chemical Society. (b) Reprinted with permission from ref. 28 Copyright 2004 American Chemical Society. (c) Reprinted with permission from ref. 32 Copyright 2004 American Chemical Society. | ||
Rida and Gijs27 showed that supraparticle structures can be retained at a well-defined position within a microchannel by focusing the magnetic field and by using ferromagnetic particles. Using an alternating magnetic field, they found that the rotational motion of the particles enhances the fluid perfusion through the magnetic structure.28 The strong particle–fluid interaction could be controlled by the field frequency and amplitude, as well as the fluid flow rate, and the mixing effectiveness was analyzed by studying two parallel flow streams within the microchannel (see Fig. 2b). Starting from a laminar flow pattern, a 95% mixing efficiency was obtained using a mixing length of only 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm and flow rates on the order of 0.5 cm s−1. The efficient fluid mixing was attributed to the chaotic splitting of fluid streams through the dynamic and randomly porous structure of the particle aggregate, combined with the relative motion of the fluid with respect to the magnetic particles. This type of magnetic micromixer was also studied using a numerical model in order to find the optimal magnetic actuation conditions for different microchannel dimensions.29 In another study,30 microchannels were connected to a microfluidic mixing chamber in which ferromagnetic particles were actuated using rotating fields to efficiently mix fluids flowing at velocities up to 5 mm s−1. Suzuki et al.31 combined a two-dimensional serpentine channel with pulsed lateral magnetic particle translation to create the typical stretching and folding behavior of fluid that is characteristic for chaotic mixing.
μm and flow rates on the order of 0.5 cm s−1. The efficient fluid mixing was attributed to the chaotic splitting of fluid streams through the dynamic and randomly porous structure of the particle aggregate, combined with the relative motion of the fluid with respect to the magnetic particles. This type of magnetic micromixer was also studied using a numerical model in order to find the optimal magnetic actuation conditions for different microchannel dimensions.29 In another study,30 microchannels were connected to a microfluidic mixing chamber in which ferromagnetic particles were actuated using rotating fields to efficiently mix fluids flowing at velocities up to 5 mm s−1. Suzuki et al.31 combined a two-dimensional serpentine channel with pulsed lateral magnetic particle translation to create the typical stretching and folding behavior of fluid that is characteristic for chaotic mixing.
Another interesting development has been the use of multiple small particle plugs instead of one large particle plug. Multiple plugs can be stably formed and retained in a channel by integrating soft-magnetic elements in the channel walls,33 or by using a channel with a periodically varying cross-section and a magnetic field orthogonal to the channel (see Fig. 2c).32 The advantage of using distributed particle plugs in a fluid flow is that the biochemical reactions on the particles are more easily controlled and monitored.
Magnetic particles are effective for achieving fluid mixing in microchannel flows, but strong forces are needed to retain the particles in the microchannel. Generally, ferromagnetic particles with high magnetic content need to be used because the magnetic forces achievable with superparamagnetic particles are too weak.34 However, Moser et al.35 showed that superparamagnetic particles can be used if the fields are generated by a combination of ferromagnets and electromagnets. An advantage of using superparamagnetic particles instead of ferromagnetic particles is that superparamagnetic particles lose their magnetization when the field is turned off, facilitating the redispersion of particles in solution. This is very important in case the particles are needed for further assay steps such as target capture or detection.
2.2 Mixing a static fluid using magnetic particles
In microfluidic systems, fluid mixing is not necessarily performed in a fluid flow. Fluid mixing can also be performed in static fluid compartments, e.g. to mix a sample with reagents after loading the sample into a microfluidic reaction chamber, or to homogenize reagents and avoid near-surface reactant depletion during a biochemical reaction at a surface. For such applications, superparamagnetic particles have been shown to be very useful, as we describe below.Vuppu et al.36,37 were one of the first to discover that superparamagnetic particles form rotating chains in a rotating magnetic field. The rotors could be assembled dynamically and the length and speed were found to be varying in time (see Fig. 3a), and as such the method was reported to be suitable for micromixing applications in biosensing.
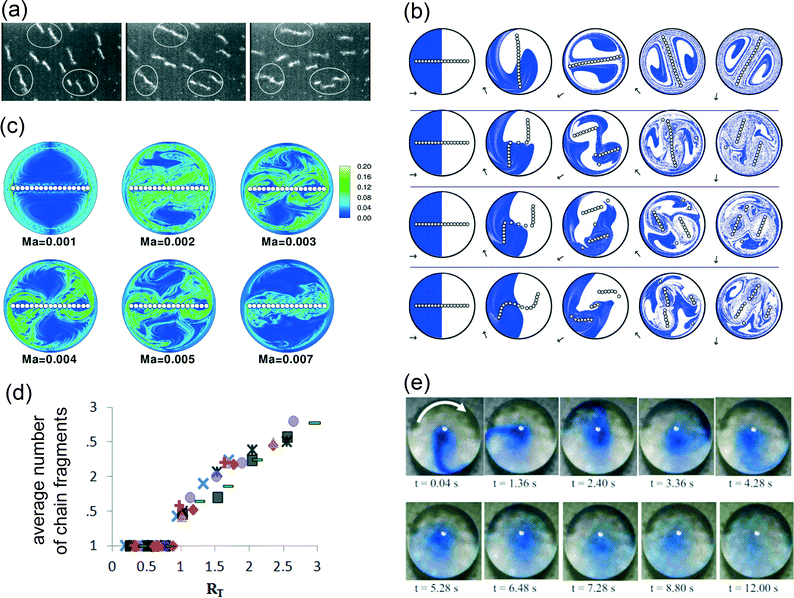 | ||
| Fig. 3 Examples of magnetic particle-based mixing within a static fluid compartment. (a) In a rotating field (3.2 Hz), magnetic particles form chains that grow and fragment dynamically. (b) Results of 2-D numerical simulations of a particle chain in a rotating field, showing the progression of mixing at different time points (to the right), for four different Mason numbers: (from top to bottom) Ma = 0.001, 0.002, 0.003 and 0.005. Initially, the fluid interface is perpendicular to the chain. (c) Results of 2-D numerical simulations, showing the spatial distributions of the maximum Lyapunov exponent at several Mason numbers. (d) The experimentally determined time-average number of chain fragments as a function of RT (as defined by eqn (2)). The external magnetic field is kept constant at 5 mT (square, diamond, triangle, circle, rectangular) and 9 mT (crosses). The experimental rotating chains were varied from N = 0 to N = 14. (e) Magnetic field-induced mixing of a dye in a microliter droplet by magnetic particles (∅2.65 μm) suspended within the fluid. (a) Reprinted with permission from ref. 36 Copyright 2003 American Chemical Society. (b, c) Reprinted with permission from ref. 17 Copyright 2007 The American Physical Society. (d) Reprinted with permission from ref. 41 Copyright 2012 The American Physical Society. (e) Reprinted with permission from ref. 43 Copyright 2009 The American Institute of Physics. | ||
To understand the dynamic behavior of the chains in more detail, models have been developed in which particle chains were treated as three-dimensional circular cylinders,38 as chains of circles in two dimensions17,39 and finally as chains of spheres in three dimensions.40–43 To characterize the chain behavior, most studies17,39,40,44 used the dimensionless Mason number, which is the ratio between the rotational shear forces (i.e. hydrodynamic drag) and the magnetic interaction forces (i.e. the magnetic forces):
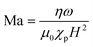 | (1) |
As the reported optimal values for the Mason number vary in the literature, a modified Mason number was introduced by Gao et al.41 which more exactly describes the acting torques and includes the number of particles, N, within a chain. The number RT equals the ratio of torques (rather than forces) and was defined as:
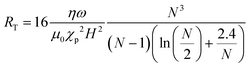 | (2) |
Experimental mixing studies were performed in microliter reaction chambers42,46–48 and in droplets43 (see Fig. 3e). In general, the experiments confirm theoretical data,47 but simulations fail to describe systems in which the particle density is high. Over time, particle chains grow in length48 and interact with other chains,43,46,47 which are not covered in simulations of isolated chains.
In conclusion, the mixing capabilities of chains of superparamagnetic particles in a rotating magnetic field have been well studied. The optimal chaotic mixing is obtained for long chains that exhibit breaking and reformation behavior. This type of mixing is particularly interesting to accelerate (bio)chemical reactions in static microfluidic compartments, as it can homogenize fluids and thereby overcome diffusion limitations.
3 Capture of analyte using magnetic particles
The high surface-to-volume ratio and the availability of many bio-functionalization options make magnetic particles well-suited for the capture of analyte from biological samples. The analyte capture can be of specific as well as non-specific nature. Non-specific capture has been mainly developed for the isolation and purification of nucleic acids from lysed samples. In particular, magnetic silica particles have been found to be very useful for nucleic acid preparation and detection.49–52 The capture process relies on the physisorption of the nucleic acids to the particles and is followed by fluid exchange steps to achieve isolation and purification. Specific capture requires the functionalization of particles with specific capture molecules, such as antibodies, with a high affinity to the analyte that is to be detected. In either case, the analyte capture rate scales with the total surface area of the suspended particles and therefore with the particle concentration. However, the use of a very high concentration of particles has disadvantages for downstream processes in an integrated multi-step lab-on-chip assay. High particle concentrations generally increase non-specific particle–particle and particle–surface interactions, enhance field-induced particle aggregation, cause steric hindrance in particle concentration steps, obstruct chemical reactions on the particles, and sterically hinder reactions between the particles and a biosensing surface. Therefore, it is desirable to decrease particle concentrations while maintaining high capture rates. To this end, magnetic actuation-based mixing techniques can be applied as discussed in the previous section. In the following paragraphs, we will discuss the effects of applying magnetic actuation for analyte capture, with the focus on the process of specific analyte capture. We describe the basic mechanisms underlying particle-based affinity capture of target analytes and review the literature on specific analyte capture by magnetic particles in flowing and in static fluids.3.1 The analyte capture process
The specific capture of analytes from a fluid onto magnetic particles is driven by (i) the encounters between target analytes and bio-functional molecules on the surfaces of the particles, and (ii) the subsequent biochemical reactions between analytes and the surface-coupled capture molecules. This creates two avenues to accelerate the capture rate, firstly by increasing encounters and secondly by increasing the biochemical reaction rate. The specificity of the capture is generated by the biochemical reaction. For example, in immunoassays, antibodies are coupled to the particles for specific capture of analytes from the fluid. The analytes are typically present in very low concentrations within a complex sample containing a high concentration of background material, such as blood or saliva. In such complex matrices, non-specific adhesion of non-targeted molecules to the magnetic particles can seriously hamper the effectiveness of the assay. Therefore, it is essential to have control over the surface properties and to have a detailed understanding of the specific and non-specific surface reactions.The process of particle-based capture of target analytes is similar to a bimolecular binding process,53,54i.e. it consists of an encounter step between the two components, which leads to a transient complex that can subsequently react chemically and become a bound complex. The total process is characterized by the overall rate constant of association, ka (unit: M−1 s−1). For typical protein–protein interactions, the association rate constant ranges between 103 and 109 M−1 s−1. The association rate constant of specific protein–protein interactions is, to a large extent, determined by the fact that the two macromolecules can only bind if their outer surfaces are aligned and oriented in a very specific manner.53 A relative translation by a few Angstroms or a relative rotation of a few degrees is enough to break the specific interactions. In general, the association rate of a protein complex is limited by diffusion and by geometric constraints of the binding sites, and may be further reduced by the final chemical reaction. In practice, usually either diffusion or the chemical reaction dominantly limits the reaction rate, although there is no simple test to determine which process is the most important. Nevertheless, there are two indications for a diffusion limitation.53 First, diffusion-controlled rate constants are usually high (>105 M−1 s−1). Second, diffusion-controlled association involves merely local conformational changes between unbound proteins and the bound complex, while reaction-controlled association typically involves gross conformational changes such as loop reorganization or domain movement.53 Typical antibody–antigen association rate constants are in the range of 105–107 M−1 s−1, which indicates that such reactions are generally diffusion-controlled.53,55,56
While most literature has focused on understanding the bimolecular reaction between two proteins that are free in solution, here we are interested in the case that one of the proteins is immobilized on the surface of a particle. So, the bimolecular reaction involves the binding between a reactive particle and a protein, where the biochemical specificity to the targeted protein is determined by the capture proteins coupled to the particle. We have performed an experimental study57 to identify to what extent the different stages of the binding process are limiting. In particular, the diffusional encounter step was split up into the process of diffusional transport through the fluid volume and the process of near-surface alignment (see Fig. 4). Where volume transport generates the first encounters between particles and target proteins, the subsequent near-surface alignment process deals with the alignment rate of the binding sites of the reactants. The volume transport is essentially a translational process, while the alignment is determined by both the translational and the rotational mobility of the reactants. The following reaction equation was used to describe the capture process of a fluorescent target nanoparticle (FT) by a magnetic capture particle (MC):
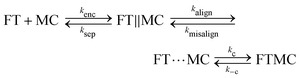 | (3) |
 | ||
| Fig. 4 Effective affinity capture of targets depends on the reactive surface properties of magnetic particles. Schematic representation of the different stages of the capture process of targets (in green) by capture particles (in gray). Targets and capture particles are sketched with multiple binding sites (in red). The stages are as follows: (left) volume transport creates encounters between magnetic particle and targets, (middle) near-surface transport creates alignment of binding sites, and (right) bonds are formed by chemical reaction. Reprinted with permission from ref. 57 Copyright 2013 American Chemical Society. | ||
When free proteins react in solution, the alignment process (i.e. rotational diffusion) is an important restriction due to the highly specific alignment constraints,55,58 but volume transport (i.e. translational diffusion) is not a limitation. In case one of the two proteins is attached to a surface, however, volume transport can become a limitation.59 Depending on the number of binding sites at the surface and the intrinsic chemical reaction rate, reactants can become depleted close to the surface, and depletion can be reduced by actively transporting the fluid over the surface.60 Therefore, depletion can also play a role in particle-based target capture, which means that reaction rates may be positively influenced by actively applying volume transport processes. Indeed, increased reaction rates have been observed when actively moving particles through the fluid using magnetic fields,61 and when applying flows to induce fluid perfusion through clusters of magnetic particles.62 Limitations due to specific alignment constraints can be further overcome by maximizing the number of binding sites on the particle surface and by improving the orientation of the immobilized proteins. Equally important is that the surface properties of the particles should be optimized to reduce non-specific interactions and to make particle-based assays suitable for operation in complex fluids. In practice, the surface optimization process is more easily performed for particles than for planar surfaces, due to the fact that surface engineering can be applied with a higher throughput to particles in solution than to surfaces in microdevices.
3.2 Analyte capture using magnetic particles in a flowing fluid
Improving the capture efficiency of target analytes from a fluid means that the interaction between the analyte and the capture agents (e.g. antibodies) should be maximized. For example, in surface plasmon resonance biosensing, a surface with immobilized antibodies is used to capture analytes from a fluid that is flowing past the surface.63 Without a flow, the analyte concentration at the surface can become limited by diffusion, which reduces the binding rate. The application of a fluid flow overcomes the diffusion limitation, maintains a maximum analyte concentration at the surface and therefore keeps the binding rate at a maximum value. The total binding rate scales linearly with the number of binding sites and therefore also with the available surface.Immobilizing antibodies on micro- or nanoparticles increases the reactive surface-to-volume ratio during incubation. In a moving fluid, the particles, in principle, follow the fluid flow such that the particle–fluid interaction is not improved unless other forces are exerted on the particles. Here, the magnetic properties of magnetic particles can be exploited, as external fields can be used to retain the particles within a fluid stream.64,65 Hayes et al.64 showed that a bed of superparamagnetic microparticles can be formed within a micro-capillary, which can be used for a flow-based immunoassay. However, the static configuration was far from optimal, as fluid perfusion through the bed was minor and most fluid flowed past the bed, requiring the bed to be 1–3 mm in length. In addition, the applied flow rates were limited (~0.1 cm s−1) in order to allow analytes to diffuse into the particle bed and to prevent superparamagnetic particles from being dragged along with the flow.
To improve flow-based assays, other groups replaced superparamagnetic particles by ferromagnetic particles, and formed dynamic particle structures over the whole cross-section of a channel (see Fig. 5a).28,66 By blocking the whole cross-section of the channel, all fluid passed through the small pores between the particles. It was shown28 that alternating the local magnetic field enhanced the perfusion of fluid through the randomly varying porous structure of the particles. To reduce the cluster size and to improve confinement of small particle clusters in plugs, microchannel structures have been modified using microfabrication techniques.32,67
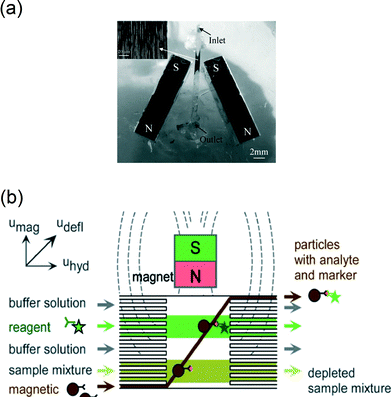 | ||
| Fig. 5 Examples of magnetic particle actuation to capture analytes from fluid flows. (a) Using two magnets near a microchannel, plugs of superparamagnetic particles can be formed and retained within the channel as shown in the inset. The plugs are used to accelerate reactions of particles with targets in a fluid that is flowing through the channel. (b) Alternatively, magnets have been used to move magnetic particles through several reagent streams in a continuous flow reactor. (a) Reproduced from ref. 66 with permission from The Royal Society of Chemistry. (b) Reproduced from ref. 69 with permission from The Royal Society of Chemistry. | ||
The use of ferromagnetic particles seriously complicates downstream detection steps, because the magnetically induced clustering of the particles is practically irreversible in a microchip. Superparamagnetic particles suffer much less from irreversible magnetic clustering, but in a flow the particles are not easily retained as they typically have smaller magnetic moments. Very strong field gradients are required35,62,68 which complicates microfluidic integration. Nonetheless, superparamagnetic particles with high magnetic content have been shown to be useful for target capture in a microfluidic flow, for example, by moving particles laterally through flow streams containing different reagents within a single microchannel, and by subsequently collecting the particles in a separate outlet (see Fig. 5b).69–73 Ganguly et al.73 found that this method of magnetophoretic mixing strongly depends on the particle concentrations, the flow fraction of the analyte stream, and the flow rate. In magnetophoretic mixing, particle–fluid interactions can be high, and the superparamagnetic nature of the particles, in principle, enables the addition of subsequent detection steps.
Overall, several studies have reported the use of actuated magnetic particles in microfluidic flows to perform analyte capture from the fluid. In most studies, analyte capture was evaluated by determining the limit of detection (LoD). However, it is difficult to attribute significance to the reported LoDs due to the lack of standardized conditions (biomarker, biomaterials, sample matrix, incubation times, detection method, data analysis, etc.) as we will discuss in section 4. It would be useful if future studies expressed the data in terms of association rate constants and included the dependence on process parameters such as flow speed, number of particles, etc. This may help to gain insight in the role of the underlying processes (such as translational and rotational transport limitations) and will help to reveal scaling relationships for the microfluidic system design.
3.3 Analyte capture using magnetic particles in a static fluid
An alternative approach to perform analyte capture from fluid is to actuate magnetic particles in an overall static fluid volume, within the so-called stationary microfluidics concept. An advantage of performing analyte capture in a non-flowing fluid is that the sample fluid is very efficiently used, because no sample is discarded in order to develop a flow. Furthermore, fluid pumping techniques are not required, which simplifies the total microfluidic system, i.e. the cartridge, the analyzer, and the cartridge–analyzer interface.Analyte capture using magnetic particles in a static microfluidic chamber was shown by Bruls et al.25 A sample fluid was inserted in a cartridge and filled the reaction chamber by capillary forces. Thereafter, a dry reagent containing magnetic particles was automatically dissolved into the fluid, allowing the particles to capture the analyte from the solution. The capture process itself was not actively steered by magnetic forces. Tanaka et al.74 showed that rotating magnetic fields (at 30–90 Hz) can be used to agitate clusters of magnetic particles in a reaction chamber in order to bind target proteins on the antibody-coated surface of the particles. Quantitative data on the enhancement of the binding rate were not shown. In another study,75 magnetic particles were suspended in a capillary tube that was positioned between two ferromagnets. Due to the magnetic fields, the particles arranged themselves into chains, and by rotating the capillary, the particle–fluid interaction was enhanced. Target capture was quantified using an ELISA and compared for different Mason numbers by altering the rotation speed of the capillary tube (see Fig. 6a). It was found that an acceleration by at least one order of magnitude could be obtained by rotating magnetic particle chains compared to a zero rotation speed of the capillary tube. It was found that small Mason numbers (corresponding to long chains that break and reform) generate the largest enhancement for analyte capture. In an inverse arrangement, Gao et al.47 applied rotating magnetic fields to magnetic particles suspended in a reaction chamber (see Fig. 6b). They found that rotating particle chains that exhibit breaking and reformation behavior (resulting in chaotic fluid mixing) enhance the capture rate of target analytes by a factor of 3 compared to rigid rotating particle chains. The breaking and reformation behavior of particle chains was enhanced by alternating the Mason number over time, i.e. by alternating between high and low frequency actuation. In this way, global mixing was alternated by local mixing. However, the Mason number is not the optimal number to describe the system, because it was defined for individual chains, not for large numbers of mutually interacting chains. Where a single isolated chain would remain rigid, a non-isolated chain at the same Mason number would grow further and maintain breaking and reformation behavior.
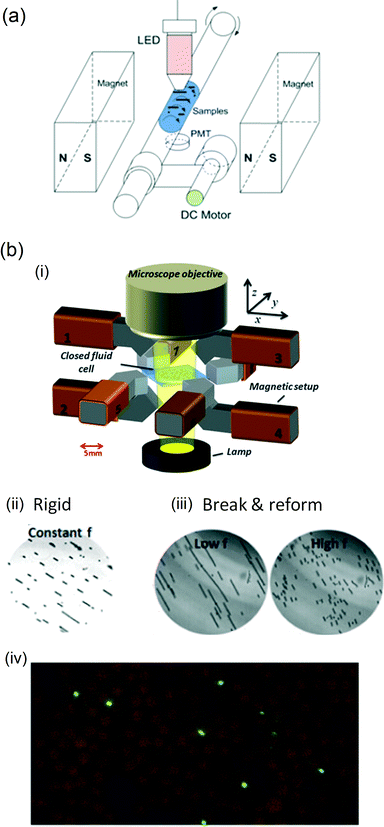 | ||
| Fig. 6 Rotation of magnetic particle chains to accelerate target capture. (a) Chain rotation induced by rotating a capillary tube in a static magnetic field. (b) Alternatively, particle chains have been rotated by applying rotating magnetic fields with (i) an 8-pole electromagnet. (ii) Using constant field rotation frequencies, particle chains are found to remain rigid during rotation; (iii) whereas by alternating high and low rotation frequencies, the so-called break-and-reformation behaviour can be induced to the particle ensembles. (iv) For both types of actuation, the effect on the capture rate of biotin-coated fluorescent nanoparticles was quantified by counting magnetic particles (∅2.8 μm; brown) and the captured targets (∅200 nm; green) in microscope images after incubation. (a) Reprinted with permission from ref. 75 Copyright 2012 American Chemical Society. (b) Reprinted and adapted with permission from ref. 47 Copyright 2013 Springer. | ||
These studies show that magnetic particle-based target capture can be accelerated by applying time-dependent magnetic fields on particles in a static fluid volume. As mentioned in the previous section, it would be useful if future studies reported data in terms of the association rates and included the influence of actuation parameters, such as field strength, field frequency and field direction. This would allow for a comparison between the different devices and actuation methods.
4 Analyte detection
After the capture of target analytes, further processing is needed for accurate and specific detection. When the magnetic particles are used only as carriers, then the captured analytes are exposed to further (bio)chemical processes and are typically detected by luminescent labels such as enzymes or fluorescent molecules. Magnetic particles can also serve as a label, to signal molecular binding at a sensing surface, or to signal molecular binding between particles in an agglutination assay. Here, we review different methods for the detection step with the focus on the potential for total lab-on-chip integration of the assay.An evaluation of the analytical performance of a detection methodology is often done by measuring dose–response curves. In many publications, attention is primarily given to the limit of detection (LoD) derived from a dose–response curve.76 However, papers of exploratory research generally report very limited statistics. The LoDs are mostly based on a low number of data points and LoD confidence intervals are hardly ever given. Furthermore, the chosen assay (biomarker, biomaterials, sample matrix, incubation times, etc.) has a strong influence on the LoD, so the LoDs of papers with different assays cannot be compared. Finally, for medical applications, it is not the LoD but the limit of quantification (LoQ) that is of prime relevance, i.e. the lowest biomarker concentration that can be quantified with a given required precision (typically <30%).77,78 The LoQ is close to the LoD if a dose–response curve has a good sensitivity, i.e. if the signal changes strongly as a function of the target concentration. However, if a dose–response curve has a very weak dependence on the concentration (e.g. if the signal scales with the logarithm of the concentration), it might even be impossible to precisely quantify the target concentration. In view of the above, when discussing the literature, we will mention the measured target and the sample matrix, the character of the reported dose–response curves, and the order of magnitude for the LoD or LoQ. When estimating the LoQ for linear dose–response curves, we will use the definition that the LoQ equals ten times the standard deviation of the blank divided by the slope of the dose–response curve.78
4.1 Magnetic particles as carriers
When magnetic particles are used as carriers or substrates for the detection of target analytes, then the particles are first used to capture target analytes, subsequently the captured analytes are labeled and finally the label is detected.7 For accurate detection, it is important that only bound analytes are labeled, and that only bound labels are detected. This requires that several washing or separation steps are performed, i.e. the particles need to be exposed to different fluids. After the labeling step, detection can be performed close to a surface or in the bulk of the fluid.As detection labels, fluorescent dyes or chemiluminescent molecules are most frequently used. Fluorescent labels have, for example, been used in microfluidic flow-based assays with supraparticle structures,64,73 magnetic particle plugs32,68 and isolated particles.69 In these assays, the single fluorescent dye molecules could not be resolved and instead the overall fluorescence from the particles was detected, e.g. using a CCD camera. The reported detection limits are typically on the order of several tens of picomolar and assays have mostly been performed in buffer.19 The dose–response curves tend to be sublinear and as a consequence the limits of quantification are substantially higher than the reported limits of detection. The detection limits are quite high due to the fact that the signals per label are weak while the background fluorescence from magnetic particles is significant. To reduce background, evanescent fields have been applied to only excite fluorescent dyes close to a substrate,80 but this approach has not resulted in a clear improvement of the detection limit.
To allow detection with single-molecule resolution, Todd et al.13 used magnetic particles to capture the analytes and thereafter labeled the analytes with fluorescent detection antibodies. After a buffer exchange, the detection antibodies were eluted, separated from the magnetic particles and counted with single molecule resolution while flowing through a capillary. From the data, we estimate a LoQ of a few femtomolar for the detection of interleukin-17 in plasma or serum. The dose–response curve shows a dynamic range of almost 5 orders of magnitude. For other analytes – among which were cardiac troponin (cTnI) and tumor necrosis factor (TNF-α) – similar or slightly higher LoQs were obtained. The method gives a very high sensitivity, but requires numerous fluid handling steps (e.g. washing, buffer exchange, elution, flows) and would be difficult to miniaturize for point-of-care testing.
Enzymes have also been used as labels in the so-called enzyme-linked immunosorbent assays (ELISAs). Enzymes that convert substrates suited for luminescence75,79,81–85 or electrochemical86,87 detection are available. Generally, luminescence-based ELISAs involve a washing step after enzyme labeling, followed by exposure of the particles to a fluid containing the reactants, after which bulk luminescence is detected.75,81–83 A main advantage of using enzymatic labels is that the signal is amplified by the enzymatic conversion process. To reach single-target sensitivity, Rissin et al.79 loaded single magnetic particles in separate microwells, which were subsequently used as isolated ELISA reaction chambers (see Fig. 7). When the number of particles is higher than the number of targets, digital counting is possible, which allows the detection of single analytes according to Poisson statistics. In buffer, streptavidin-β-galactosidase could be detected with an estimated LoQ of a few attomolar and a dynamic range of 4 orders of magnitude. In serum, prostate specific antigen (PSA) and TNF-α could be detected with estimated LoQs close to a femtomolar (based on the quasi-linear dose–response curve at low concentrations). Concerning the integration of the technology, several aspects have been investigated such as rapid array loading,88 low-cost fabrication of array wells84 and multiplexed assays,89 but complete miniaturization and integration are still a challenge.
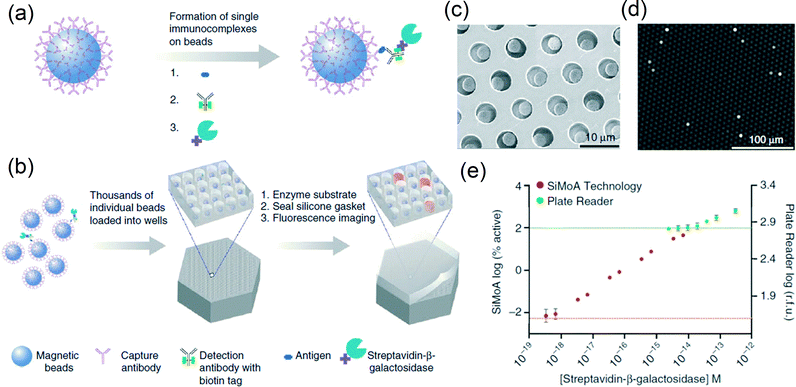 | ||
| Fig. 7 Digital ELISA based on arrays of femtoliter-sized wells. (a, b) Single protein molecules were captured and labeled on magnetic particles (∅2.8 μm) using standard ELISA reagents (a), and particles with or without a labeled immunoconjugate were loaded into femtoliter-volume well arrays for isolation and detection of single molecules by fluorescence imaging (b). (c) Scanning electron micrograph of a small section of a femtoliter-volume well array after particle loading. (d) Fluorescence image of a small section of the femtoliter-volume well array after signals from single enzymes are generated. The concentration of protein in bulk solution is correlated to the percentage of particles that carry a protein molecule. (e) The log–log plot of the signal output (% active particles for the single-molecule array (SiMoA) or relative fluorescence units (r.f.u.) for the plate reader) as a function of the concentration of streptavidin-β-galactosidase (SβG) captured on biotinylated particles. Reprinted with permission from ref. 79 Copyright 2010 Nature Publishing Group. | ||
Another assay employing magnetic particles is the so-called nanoparticle-based bio-bar code assay described by Nam et al.9 Analyte targets were captured by magnetic microparticles and subsequently labeled with gold nanoparticle probes that contained both specific antibodies and DNA fragments. Magnetic separation and buffer exchange were performed, and thereafter the DNA was amplified and stained for optical detection. For detection of PSA in buffer, we estimate a LoQ of a few attomolar from the reported dose–response curve. The method gives a very high sensitivity, but the numerous fluid handling steps strongly complicate the integration into a point-of-care system.
Hahn et al.90 demonstrated an assay in which relatively large magnetic particles with a diameter of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm were labeled with magnetic nanoparticles. The labeled particles were detected by isomagnetophoresis, which discriminates small differences in magnetic susceptibility by measuring particle deflection in a streaming paramagnetic salt solution. The measurement principle is based on a delicate balance between magnetic and fluidic forces, and would be difficult to integrate into a point-of-care system.
μm were labeled with magnetic nanoparticles. The labeled particles were detected by isomagnetophoresis, which discriminates small differences in magnetic susceptibility by measuring particle deflection in a streaming paramagnetic salt solution. The measurement principle is based on a delicate balance between magnetic and fluidic forces, and would be difficult to integrate into a point-of-care system.
The above-mentioned assays involve the application of microfluidic fluid flows and/or pipetting steps. An alternative to manipulating fluids is to keep the fluids stationary, and to transport magnetic particles from one stationary fluid to another stationary fluid by using magnetic forces.23,24,50,92–95 The two fluids, e.g. fluid A and fluid B, are separated by a medium that does not mix with the aqueous fluids, for example a non-polar fluid or a gas. When a sufficiently high force is applied to the magnetic particles, the particles are pulled out of fluid A through the interface between fluid A and the medium; thereafter they are pulled through the interface between the medium and fluid B, and into fluid B. Alternatively, the fluid interfaces can be moved over the particles, e.g. by using electrowetting.82,96 The process of transferring magnetic particles between the fluids is controlled by a balance between the magnetic forces on the particles and the forces caused by interfacial tension, so-called capillary forces. Therefore, we propose to very generally refer to these transfer mechanisms as Magneto-Capillary Particle Transfer (MCPT).
MCPT has been studied in many different device geometries, like tubes,97,98 single-plane devices,23,24,51,82,94,96 vertical slits,93,99 arrays of wells92 and bi-plane capillary devices.50,91,95,100,101 MCPT has been applied for the purification and enrichment of nucleic acids23,24,50,91–93,95,98,102 and proteins.50,97,100 For example, den Dulk et al.50 used a bi-plane capillary device with liquid–gas interfaces and reported enrichment factors for nucleic acids and proteins between one and two orders of magnitude. Furthermore, it was shown that high washing efficiencies could be obtained due to very low amounts of co-transported fluid. Using MCPT, complete lab-on-chip assay integration has been demonstrated including the detection step. Nucleic acids51,101 and proteins91 have been detected, using oil-filled51,101 or air-filled91 capillary valves. The latter paper reported assay dose–response curves. Magnetic particles were moved over a cartridge through different chambers and incubated in each chamber under actuation by a rotating magnetic field (see Fig. 8). In the assay, target interleukin-8 in buffer with 10% human serum was captured and fluorescently labeled after a washing step. The corresponding dose–response curves had a segment with a linear relationship between signal and concentration, with the LoD and LoQ in the picomolar range.
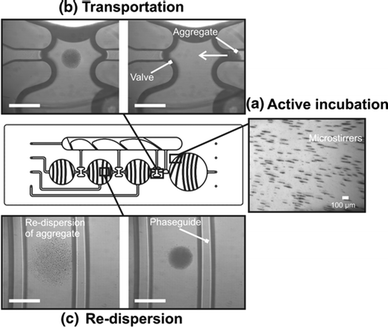 | ||
| Fig. 8 Example of Magneto-Capillary Particle Transfer (MCPT) in a carrier-only microfluidic assay in which the detection step has been integrated. In the center of the figure, the microfluidic cartridge is sketched containing several fluid chambers and valves to enable magnetic particle transfer between different fluids. The bright-field images show the different types of magnetic actuation that were applied to the magnetic particles: (a) active incubation via the assembly of particles into magnetically rotating microstirrers (oriented parallel to the chip plane); (b) particle transportation from chamber to chamber, using a focused magnetic field; and (c) re-dispersion of the aggregate by moving the focused magnetic field downwards and away from the chip, thus causing the aggregate to be pulled apart. The scale bar is 1 mm and the white arrow shows the direction of transport. Reprinted from ref. 91 with kind permission from Springer Science + Business Media. | ||
In the carrier-only assays, the particles are exposed to multiple fluids, as is also done in high-throughput robotic pipetting-based assays. Carrier-only assays have the advantage that very similar reagents can be used as in commercial pipetting-based assays. Another advantage is that the detection can occur in the bulk fluid, so the control of cartridge surfaces is not very critical. The most important disadvantage is that the integration of multiple fluids complicates the device technology. Furthermore, carrier-only assays require relatively strong magnetic fields, which can cause non-specific binding between the particles. For reliable detection, particle aggregation needs to be overcome, and we will further discuss this in section 5.
4.2 Agglutination assays with magnetic particles
Agglutination assays exploit a process wherein aggregates of particles are formed when specific analytes are present in the sample fluid.‡ The particles are provided with target-binding molecules and the analytes should have at least two epitopes that can react with the particles. The degree of aggregation is a measure for the concentration of analytes within the fluid. In magnetic agglutination assays, the formation of particle clusters is accelerated by bringing particles together under the influence of a magnetic field. Two types of magnetic agglutination assays can be distinguished: (i) assays in which the magnetically actuated particles form clusters while being exposed to a fluid stream, and (ii) assays in which the fluid is static.An assay with a streaming fluid was reported by Degre et al.103 Magnetic particles were flowed through a microfluidic channel past two external magnets. The combination of the attractive magnetic dipole–dipole interactions between particles and the shear flow results in the formation of chain-like particle clusters. Beyond the magnets, clusters disaggregate or remain clustered depending on the number of captured analytes. In the original publication,103 no dose–response curves, high-statistics data, or measurements of noise or non-specific background were reported.
Moser et al.62 applied localized magnetic fields to retain and dynamically actuate superparamagnetic particles in a microchannel flow, and thereby enhanced the perfusion of the magnetic particles (see Fig. 9a). After switching off the magnetic field, the thermal diffusivity of the particle cloud was measured, revealing the degree of particle aggregation. The area of the released plug was measured to quantify analyte capture. A dose–reponse curve was reported for the detection of biotinylated bovine serum albumin (BSA) in buffer, showing a logarithmic characteristic. The LoD is in the picomolar range, but the LoQ will be much higher due to the logarithmic dose–reponse characteristic.
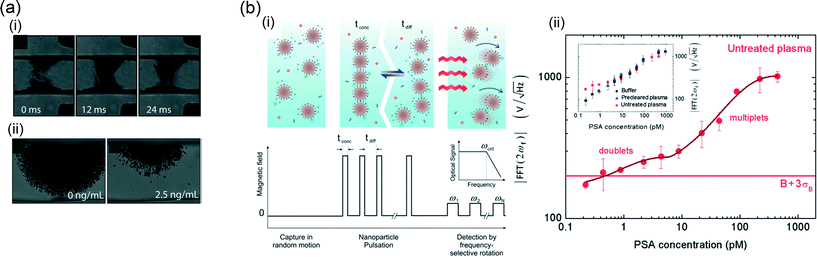 | ||
| Fig. 9 Agglutination sandwich assays based on magnetic particles. (a,i) A dynamic particle plug in a microchannel with a flowing fluid. The particles were actuated by an external magnet at 70 Hz. The continuous motion of the particles in the center of the channel allows efficient perfusion by the analyte solution. (a,ii) The amount of agglutination depends on the target concentration and was determined by analyzing the area of the released plug after turning off the external field. A streptavidin-bBSA model system was used. (b) One-step homogeneous assay technology based on magnetic nanoparticles in a static fluid. (b,i) First, the biological sample was spiked with nanoparticles. Targets were captured by diffusive motion. Subsequently, cluster formation was accelerated by applying magnetic fields in pulses to bring particles together (tconc) and to allow diffusion to enhance bond formation (tdiff). The formed clusters were detected by applying rotating fields with increasing rotation frequencies and by measuring the optical scattering signal. The result was a curve of the optical scattering signal as a function of the frequency (see the inset); the plateau reveals the number of clusters in solution, while the critical frequency reveals the cluster size and the viscosity of the sample. In (b,ii), the resulting dose–response curve for PSA in untreated blood plasma is shown. The inset shows the dose–response curves in buffer, precleared plasma, and untreated plasma. The data are fitted based on a model description which includes the cluster size. The horizontal line shows the level of the blank plus three times the standard deviation of the blank. (a) Reproduced from ref. 62 with permission from The Royal Society of Chemistry. (b) Reprinted with permission from ref. 104, Copyright 2012 American Chemical Society. | ||
Baudry et al.105 showed that particle aggregates can be formed effectively in a static fluid. After target capture, field-induced chains of particles were formed in order to accelerate the formation of target-induced clusters. The particle clustering was detected using turbidimetry, as the scattering properties of particle clusters differ from unclustered particles. A slightly sub-linear dose–response curve was shown for the detection of ovalbumin in buffer, with a LoD in the low picomolar range.
Park et al.106–108 monitored the growth of particle chains in a rotating magnetic field by measuring light transmittance through the sample volume. As particles form aggregates, longer chains were obtained for increased target concentrations, causing larger fluctuations in the transmitted light. For avidin in buffer, sub-linear dose–response curves were obtained with a LoD just below the nanomolar.
Magnetic particle aggregation can also be quantified by NMR sensing, as demonstrated in many publications from the Weissleder group.109,110 The aggregation assays have nearly all been done using small nanoparticles (with a diameter of a few tens of nm) and in one publication also using microparticles.110 In the latter case, magnetic fields were applied to enhance particle aggregation. In buffer, antibodies to influenza were detected. The dose–response data were recorded on a logarithmic scale, which makes it difficult to quantify the LoD or LoQ, but picomolar target concentrations were resolvable.
When the analyte concentration is much smaller than the magnetic particle concentration, only few particle aggregates are formed, governed by Poisson statistics. Many particles will not form any clusters, some particles will form two-particle clusters, and larger clusters will be rare. Ranzoni et al.104 showed that specific doublet formation in the low-concentration regime can be enhanced by applying a pulsed magnetic field during incubation, i.e. to alternatingly bring particles in close contact and let them freely diffuse to form specific bonds (see Fig. 9b). Furthermore, the optical detection sensitivity of doublets was improved by measuring the optical scattering in a rotating magnetic field.111 A dose–response curve was shown for the detection of PSA directly in undiluted blood plasma. The curves had an undulating character (see Fig. 9b-ii), revealing regimes of clusters of different sizes, with a LoD around a picomolar.
The two main challenges of particle-based cluster assays are (i) to ensure good contact between the particles in order to increase the assay kinetics, and (ii) to minimize non-specific particle clustering in complex biological samples. Magnetic fields help to bring the particles together and thereby enhance the inter-particle binding kinetics. However, magnetic fields may also increase the non-specific binding between the particles. Assays in a fluid stream require high magnetic forces in order to avoid that the particles are pulled along with the flow. High magnetic forces increase the risk of non-specific binding between the particles. In contrast, the agglutination assays in a non-flowing fluid can be carried out in weak magnetic fields, thereby avoiding strong interactions between the particles.104 When further technological improvements are made, we expect that static-fluid magnetic agglutination assays may become a very interesting format for point-of-care applications.
4.3 Surface binding assay with magnetic particles as labels
In a surface binding assay, magnetic particles are used as labels that bind in a biologically specific manner to a surface and thereby report the presence of a specific molecular species. Most commonly a sandwich format is used, with specific binders being immobilized on the particles and on the surface, which bind the target on two different epitopes. Preferably, all analytes in the fluid become sandwiched between a particle and the surface, which is possible when the concentration of particles exceeds the analyte concentration. To ensure efficient capturing and labeling, the magnetic particles need to efficiently (i) capture targets from solution (cf. section 3), (ii) be brought to the surface, and (iii) interact with the surface on molecular length scales. The transportation toward the surface can be achieved relatively easily by applying magnetic field gradients towards the sensor surface.113,114 It is more difficult to control the particle–surface interaction, because particles concentrated at a surface mutually exhibit magnetic particle–particle and steric particle–particle interactions. In addition, rotational exposure of the particles to the surface is important and non-specific interactions between the particles and the surface should be minimized.115,116Several methods have been developed to optimize the particle–surface interactions. Bruls et al.25 developed an actuation protocol in which in-plane fields, out-of-plane fields, and field-free phases are alternated. In-plane fields bring particles to the surface, out-of-plane fields generate chains, while the field-free phase allows free Brownian motion of the particles in order to optimize their (rotational) exposure to the surface. In this way, effective specific sandwich formation was shown. The protocol keeps the particles in constant motion relative to the sensor surface, which may also minimize non-specific binding with the surface. Dose–response curves were determined for the detection of cardiac troponin (cTnI) in buffer and in undiluted blood plasma. The dose–response curves were practically linear with the LoDs around a picomolar.
Gao and van Reenen et al.112 developed an actuation protocol to induce repulsive magnetic dipole–dipole interactions between particles at a surface. The method consists of aligning particle aggregates with a surface by using field gradients and in-plane oriented magnetic fields, followed by the application of an out-of-plane magnetic field while a field gradient maintains the particles at the surface. In this way, clusters of microparticles were shown to disaggregate (see Fig. 10). By repeatedly applying these two steps, clusters consisting of tens of particles could be almost completely redispersed over the surface in several tens of seconds. Evaluation of this method in a surface binding assay has however not yet been performed.
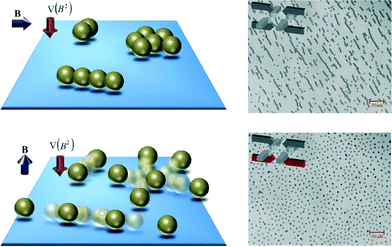 | ||
| Fig. 10 Controlling particle behavior at a sensor surface. Schematic representation and experimental data of the disaggregation of magnetic particle clusters. Clustered particles that are present in solution are drawn to a physical surface by means of a field gradient. A horizontal magnetic field is applied to align clusters parallel to the surface (top figures). Subsequent application of a vertical magnetic field breaks the particle clusters by inducing repulsive dipolar magnetic interactions (bottom figures). Reproduced from ref. 112 with permission from The Royal Society of Chemistry. | ||
Some papers report studies in which fluid flow is an essential component. Morozov et al.117,118 combined flows past a sensor surface with electrophoresis and magnetophoresis to respectively bind bacterial toxins to a surface and thereafter attract magnetic particles to the surface to form the sandwich. Shear flows were applied to wash away particles bound to the surface by weaker non-specific bonds.119 Assays were performed on five different bacterial toxins in different media (i.e. buffer, water, milk and meat extract). The reported dose–response curves all had a logarithmic character. LoDs were reported of several tens of femtomolars, however, in view of the large error bars and the logarithmic dose–response curve, the LoQs will be much higher.
Tekin et al.120 flowed magnetic particles (∅2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm) past a sensor surface that was provided with regularly spaced smaller magnetic particles (∅1.0
μm) past a sensor surface that was provided with regularly spaced smaller magnetic particles (∅1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm) coated with antibodies. In this way, the larger magnetic particles containing captured analytes were transported to the sensor surface and could interact with the smaller magnetic particles, assisted by attractive magnetic dipole–dipole interactions. Applying this method to a sandwich assay for biotinylated anti-streptavidin and TNF-α in fetal bovine serum, dose–response curves were obtained with a logarithmic character: a 100 million-fold increase in the analyte concentration gave a 5 to 10 fold increase of signal. The LoDs were reported to be of several tens of attomolar, however, due to the logarithmic dose–response curve one cannot define the LoQ, because across the whole measurement range, it is not possible to quantify the analyte concentration with an accuracy of 30%.
μm) coated with antibodies. In this way, the larger magnetic particles containing captured analytes were transported to the sensor surface and could interact with the smaller magnetic particles, assisted by attractive magnetic dipole–dipole interactions. Applying this method to a sandwich assay for biotinylated anti-streptavidin and TNF-α in fetal bovine serum, dose–response curves were obtained with a logarithmic character: a 100 million-fold increase in the analyte concentration gave a 5 to 10 fold increase of signal. The LoDs were reported to be of several tens of attomolar, however, due to the logarithmic dose–response curve one cannot define the LoQ, because across the whole measurement range, it is not possible to quantify the analyte concentration with an accuracy of 30%.
As described above, the method for assisting the binding of particles at the sensor surface is very important for the character of the dose–response curve and the resulting detection sensitivity. Now, we will discuss the next step, namely, the detection of bound particle labels at a sensing surface. A method for the detection of particle labels should be sensitive, but in addition, one should consider the influence of the different methods on lab-on-chip integration, the cost effectiveness of the resulting disposable cartridge, and the miniaturization potential of the reader instrument. In particular, one should consider the compatibility of the detection methods with the presence of magnets around the microfluidic reaction chamber and near the sensing surface, in order to allow magnetic control of the particle-based assays.
In early reports, magnetic nanoparticle labels were detected by magnetic coils,121 SQUID,122 magnetoresistive sensors11,113,123–125 and Hall sensors.126 Although it is possible to combine magnetic sensing with the application of magnetic fields,113,124 it is complicated because magnetic fields tend to perturb the measurements. Furthermore, the use of lithographically made sensing chips adds costs to the disposable cartridge and demands cartridge assembly technologies suited for high numbers of electrical interconnects.
Optical detection methods are not perturbed by the presence of magnetic fields and are compatible with cost-effective mass-manufactured cartridges. Magnetic particles can be optically detected on a transparent surface in several ways, e.g. by using bright-field illumination116,127 or dark-field illumination.117 A particular challenge is to design the system in such a way that magnet poles can be positioned very close to the sensing surface. Bruls et al.25 described a detection system based on the principle of frustrated total internal reflection (f-TIR) as depicted in Fig. 11. A light-emitting diode was used to create an evanescent wave at the sensor surface via total internal reflection. The presence of magnetic particles at the surface frustrates the evanescent wave, causing a reduction of reflected light. The amount of particles at the sensing surface was recorded as a function of time by monitoring the reflected light intensity. The advantage of using f-TIR is that it is highly surface-sensitive and suited for close integration of electromagnets.
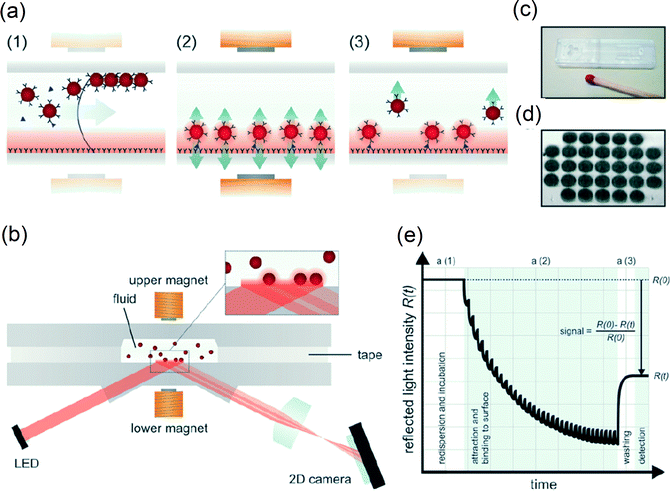 | ||
| Fig. 11 Optomagnetic immuno-biosensor based on actuated magnetic particles (∅500 nm) from the study by Bruls et al. (2009). (a) A schematic representation of the reaction microchamber showing the successive assay processes: (a1) filling of the microchamber, nanoparticle redispersion, and capturing of analyte; (a2) actuation of the particles during the process of binding to the surface; and (a3) removal of free and weakly bound nanoparticles from the sensing surface by magnetic forces. (b) The fluid microchamber placed in the optomagnetic system with electromagnets and detection optics. Light reflects from the sensor surface with an intensity that depends on the concentration of nanoparticles at the sensor surface, by the mechanism of frustrated total internal reflection (f-TIR). (c) A picture of an assembled disposable cartridge (1 cm × 4 cm) consisting of two structured plastic parts connected by double-sided adhesive tape. The cartridge contains a sample inlet, a channel, a reaction microchamber (1 μL), and a vent. (d) The f-TIR image of the magnetic nanoparticles bound to the sensor surface via an immunoassay on 31 capture spots of 125 μm diameter each. (e) A schematic real-time curve of the measured optical signal for a single capture spot. The assay phases a1–a3 are indicated. The signal modulation in phase a2 is caused by switched actuation of the magnetic nanoparticles. Reproduced from ref. 25 with permission from The Royal Society of Chemistry. | ||
Magnetic nanoparticle labels have also been detected by grating-coupled surface plasmon resonance (GC-SPR),128 which is an evanescent-field technique based on a thin gold film at the sensing surface. Different assay formats were compared, as shown in Fig. 12, for the detection of β human chorionic gonadotropin (βhCG) in buffer. Magnetic actuation clearly had a positive effect on the obtained dose–response curve, and led to detection limits within the picomolar range.
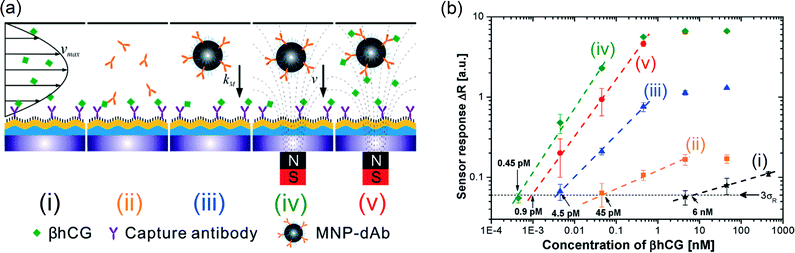 | ||
| Fig. 12 (a) Schematics of used grating-coupled SPR detection formats: (i) direct detection, (ii) sandwich assays with amplification by the detection antibody and magnetic nanoparticles coated with detection antibodies (MNP-dAB) (iii) without and (iv) with an applied magnetic field. (v) The detection format consisting of preincubating MNP-dAb with βhCG followed by the sandwich assay upon an applied magnetic field gradient. (b) The dose–response curves for the detection of βhCG by the direct detection format (i, stars), followed by antibody amplification (ii, squares) and the amplification by antibody-labeled magnetic nanoparticles without (iii, triangles) and with (iv, diamonds) an external magnetic field. In format v, a sample with βhCG was incubated with antibody-labeled magnetic nanoparticles, followed by the detection of the complexes with an external magnetic field applied (circles). Reprinted with permission from ref. 128 Copyright 2011 American Chemical Society. | ||
4.4 Magnetic stringency
A stringency process aims to improve the specificity of detection by separating unbound and weakly bound from strongly bound species. In the detection methods with particle labels discussed above, signals are generated by bonds formed between magnetic particles (agglutination assay) or between particles and a surface (surface binding assay). The bonds should be biologically specific. However, bonds can also have a non-specific nature, i.e. the bond is not mediated by an analyte molecule, which results in a false positive signal. Non-specific bonds can originate from several types of interactions, e.g. van der Waals interactions, electrostatic interactions, hydrophobic interactions, etc.129 In diagnostic tests, non-specific interactions cause background levels as well as statistical variations of the results, and thus affect the limit of quantification and the precision.The development of a diagnostic test always involves a series of optimizations in the biochemical and chemical domains in order to improve the specificity of detection, e.g. by optimizing the affinity molecules and coupling chemistries, by blocking the surfaces, and by dissolving reagents into the sample, such as pH buffers, salts, surfactants, and blockers for specific macromolecular interferences. The aim of the optimizations is to reduce the formation of non-specific bonds and to preserve or improve the specificity of the targeted bonds. In an assay involving magnetic particles, there is an additional degree of freedom, namely the forces that can be applied to the particles. Magnetic forces can be used to separate bound from unbound particles. Furthermore, when the particle labels are bound to a surface, magnetic field gradients can be applied in order to impose stringency on the bonds and thereby dissociate weak non-specific bonds. Also, the response of molecular bonds to applied stresses can be recorded, giving even more detailed information about the bonds.
An early report on the application of magnetic stringency to non-covalent bonds was published by Danilowicz et al.130 A permanent magnet was used to apply a constant force to the ensembles of bound particles and the dissociation of bonds was recorded as a function of time. Jacob et al.131 used an electromagnet which allowed a wider range of forces to be studied, thereby yielding reliable dissociation rate constants for the biomolecular bonds. It was demonstrated that populations of specific and non-specific bonds could be distinguished by the shape of the force-induced dissociation curves. In these studies, relatively large magnetic particles were used as labels (4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm in ref. 130 and 2.8
μm in ref. 130 and 2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm in ref. 131). It is convenient to use large particles for biophysical studies because large forces can be applied to single particles. However, large particles are less suited for integrated biosensing because they diffuse slowly, sediment easily, and limit the dynamic range of detection due to steric hindrance.
μm in ref. 131). It is convenient to use large particles for biophysical studies because large forces can be applied to single particles. However, large particles are less suited for integrated biosensing because they diffuse slowly, sediment easily, and limit the dynamic range of detection due to steric hindrance.
Using particles with diameters of a few hundred nanometers, Bruls et al.25,113,124 demonstrated the use of magnetic stringency in integrated surface-binding assays, see Fig. 11a, e. Here, magnetic stringency removes unbound and weakly bound particles from the surface. In fact, it replaces the fluidic wash step as found in the traditional affinity assays. The magnetic stringency obviates the need for fluid manipulation, which simplifies the assay and makes it highly suited for integration in a completely stationary assay concept.
In the future, magnetic stringency may go beyond the application of bound-free separation and the measurement of dissociation properties of molecular bonds. For example, by applying rotating magnetic fields, it has been shown that it is possible to probe the properties of DNA132,133 and protein complexes134 that are sandwiched between particles and a surface. Although still very remote from integrated biosensing, the principle of characterizing molecular bonds in a detailed biophysical manner may in the future help to further increase the specificity of biosensing.
5 Integration of magnetic actuation processes
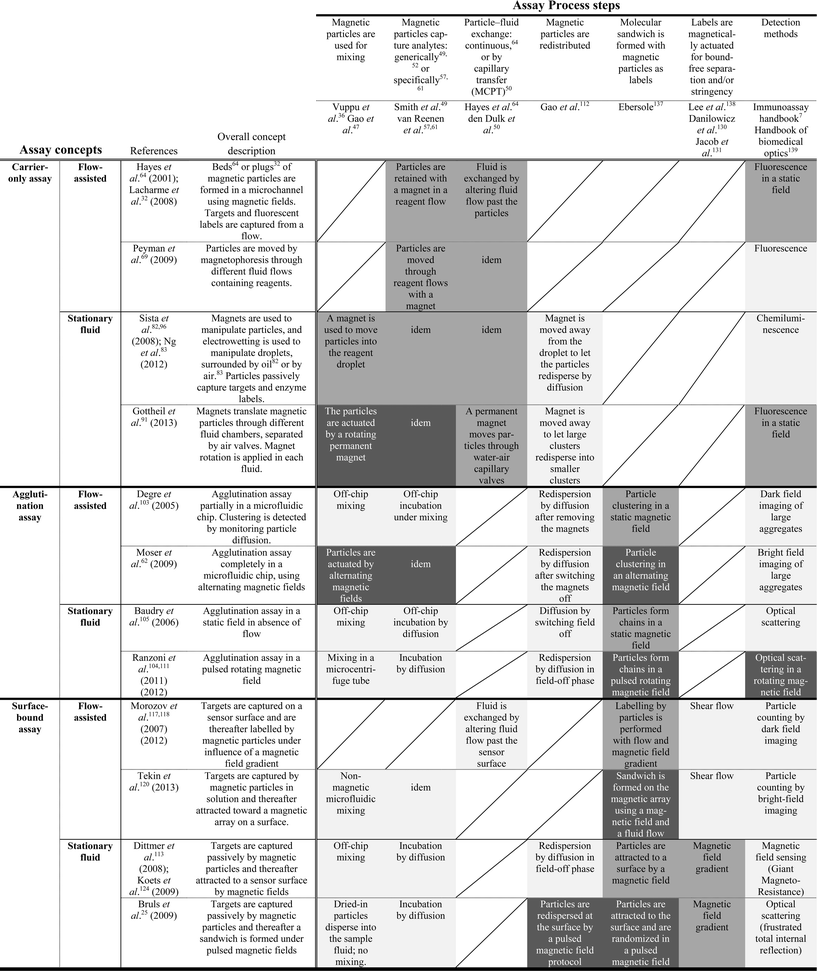
Integration is the act of making something into a whole by bringing all parts together. For an engineer, it is the process of (i) defining an overall technological function that needs to be realized, (ii) designing a system architecture and its underlying components, and (iii) quantifying all interactions within the system and feeding this back on the technical function definition. For a given functional aim, one can select different system architectures each having its own inherent advantages and challenges. As reviewed in the previous paragraphs, the manipulation of magnetic particles by magnetic fields allows one to control in a microfluidic format a list of important assay steps for diagnostic testing. Now, we will review how the integration of different assay process steps is proceeding, moving toward integrated assays that perform a series of sophisticated steps, controlled by magnetic forces.In Table 1, we have summarized the state-of-the-art use of magnetic particle actuation for integrated detection assays. The top row lists the key assay process steps. The left-most column lists the assay concepts, i.e. carrier-only assays, agglutination assays, and surface-binding assays. We made a distinction between flow-assisted devices in which active channel flows are used, and stationary-fluidic devices in which continuous fluid motion is absent. Within the table matrix, we have classified the type of magnetic actuation used at the intersection between the assay concept and the process step. The gray-scales indicate the type of actuation used: without magnetic fields (light grey), with static fields (mid grey), with dynamic fields (dark grey), or not applicable (/). The references of the process steps and the assay concepts serve as examples. The assay-concept references are focused on total assay integration; they report dose–response curves acquired by detection on the microfluidic chip.
 performed without magnetic fields;
performed without magnetic fields;  performed by applying static magnetic fields;
performed by applying static magnetic fields;  performed by applying dynamic magnetic fields; / not applied in the concept. The mentioned references serve as examples to illustrate the progress in the field; the reference list is not exhaustive. Assay-concept references have integrated the detection step and reported dose–response curves
performed by applying dynamic magnetic fields; / not applied in the concept. The mentioned references serve as examples to illustrate the progress in the field; the reference list is not exhaustive. Assay-concept references have integrated the detection step and reported dose–response curves
The first set of rows describes assays in which magnetic particles act as carriers only. In such assays the labeling step involves the addition of, for example, enzymatic or fluorescent labels. Therefore, a fluidic washing step or a fluid exchange step is essential prior to and after the labeling, in order to effectuate a separation between bound and free analytes, and between bound and free labels. One way to achieve a particle–fluid exchange step is by applying a continuous fluid flow while external magnets retain the magnetic particles within a liquid.32,64,69 Also, one can manipulate fluids by merging and splitting droplets in so-called digital microfluidics.82,83,96 Alternatively, rather than moving fluids, one can switch the magnetic particles from one stationary fluid to another, by so-called Magneto-Capillary Particle Transfer (MCPT).23,24,50,51,82,83,91–93,95,97,101 Intrinsic to carrier-only assays is the need for relatively strong magnetic fields to retain particles in a flow or to traverse capillary interfaces, with the disadvantage that particles become highly concentrated and non-specific interactions are promoted. In the mentioned papers, redistribution of particles has been effectuated by removing the magnet from the sample chamber, allowing particles to spread by diffusion. The images (see e.g.ref. 51,91) show that particle redispersion is incomplete; large clusters break up into small clusters, but not separate particles. We expect that improved disaggregation of particles, e.g. by magnetic field actuation112 and/or by further reducing non-specific interactions, will improve the assays. Magnetic stringency is not applicable in carrier-only assays because the particles are not used as labels. The carrier-only methods allow miniaturization and integration, however, a series of fluids is always needed, including active control of fluid flow and/or methods for magnetocapillary particle switching. Generally, stationary microfluidic approaches reduce the total system complexity compared to methods that require continuous flow generation.
The second set of rows describes assays based on analyte-mediated agglutination of magnetic particles. In agglutination assays, the analyte capturing and labeling steps are performed in one and the same solution. Magnetic agglutination assays have been studied in flowing fluids62,103 and stationary fluids.104,105,111 Assays in flow require higher magnetic forces, which promote non-specific binding between the particles. The highest assay sensitivities have been demonstrated with stationary-fluid agglutination assays104,105,111 using lower particle concentrations and less strong magnetic fields than used in the flow-based assays. The application of pulsed magnetic fields has been shown to increase the effectiveness of molecular sandwich formation between the particles. The increase has been attributed to a combination of two effects, namely, on the one hand, keeping the particles in close contact with each other, while, on the other hand, allowing free Brownian rotation to expose all sides of the particles. Stationary-fluid magnetic agglutination assays are highly suited for miniaturization and integration, because in principle the assays can be performed in one chamber. Thus far, magnetic field-enhanced mixing and target capture have not been applied in stationary-fluid agglutination assays, but several actuation methods47,61 seem to be suited. Furthermore, when sample pretreatment steps are desired such as analyte purification or enrichment, MCPT50 may in principle be combined with the agglutination assay.
The last set of rows in Table 1 describes the surface-bound assays, wherein magnetic particles interact with a sensor surface and form analyte-mediated bonds. The table lists assays in which a magnetic field gradient is used to attract magnetic particles to the surface, thereby enhancing the local particle concentration and promoting particle–surface binding. Magnetic field gradients have been combined with fluidic shear flows, in order to move particles past the surface and/or apply stringency to bound particles.117–120 In these papers, the surface binding steps were performed in static magnetic fields. It is important to realize that superparamagnetic particles are known to contain small ferromagnetic moments.135 Therefore, in a static magnetic field the rotation of the magnetic particles may be constrained, which may limit the surface binding effectiveness. Surface-bound assays have also been demonstrated without a fluid flow. In such stationary-fluid assays, magnetic fields have been applied to bring and keep particles near the surface25,113,124 and to randomize particle distributions.25 Using current-controlled electromagnets, the particle–surface interaction has been optimized by combining pulsed magnetic fields with field-free phases for Brownian rotation.25 Furthermore, stringency25,113,124 has been applied by reversing the field gradient. Surface-bound magnetic particle assays are highly suited for miniaturization and integration, as fluid manipulation is not necessary and the assay can be completely controlled by magnetic fields. A disadvantage of surface-bound assays is that the sensor surface needs to be biofunctionalized, which adds complexity to the assay. Important advantages are that magnetic stringency can be applied in the assay, and multiplexing can be realized by preparing binding spots with different biochemical compositions. We foresee several avenues to further control and optimize surface-bound assays by magnetic actuation. For example, actuated mixing and capture47,61 may help to further increase the speed and effectiveness of the capture process, and magnetic fields may be used to redistribute particles in the assay chamber.112
6 Conclusions
We have reviewed the use of magnetic particles and magnetic fields to perform key process steps in integrated microfluidic assays for lab-on-a-chip diagnostic applications. Magnetic particles have been applied to achieve mixing, washing and buffer exchange, both in fluid flow and in stationary microfluidic device architectures. Due to the high surface-to-volume ratio and their adaptable surface (bio-)functionalizations, magnetic particles are effective at achieving rapid and specific capture and labeling of targets. In addition, magnetic particles can be actuated for magnetic stringency steps, and can be accurately detected in complex fluids, most commonly by optical methods.Current quantitative lab-on-chip biosensing systems consist of a disposable cartridge and a reusable analyzer instrument. Cartridges are single-use objects for reasons of biochemical irreversibility and bio-safety. Therefore, it is important that a system architecture is chosen which limits the complexity of the cartridge. The development of a biosensing cartridge presents challenges in the domains of device technology (e.g. fluidics and detection) and biochemistry (e.g. reagents and bio-functionalization), and the challenges depend on the architectural choices. Broadly speaking, in the carrier-only concepts, the reagents can be close to the ones developed for pipetting-based assays; however, multiple fluids need to be controlled in the cartridge, which complicates the device technology. The agglutination assays are simpler in terms of device technology, but are demanding on the reagents because the assays are performed in one step without separation or stringency. The surface-bound assays allow sensitive particle detection and stringency, yet require careful control of the surface bio-functionalization.
The choice of a cartridge architecture is also determined by the type of assay. For some assays, it is essential to include a purification step, as is, for example, the case for most nucleic-acid detection assays. A purification step can only be performed within a multi-fluid cartridge concept. Other assays, such as sandwich immunoassays, can be performed in a single step without fluid exchange, which strongly simplifies the cartridge design.
Magnetic actuation is an enabler for lab-on-chip integration because it allows a large diversity of sophisticated fluidic and molecular process steps to be controlled by means of externally generated fields, which can strongly simplify the cartridge design. In terms of microfluidics, stationary-fluidic concepts are very attractive because they do not require continuous fluid actuation to be integrated in the system. Technologies based on continuous fluid actuation generally require large fluid volumes or complex cartridge architectures. Stationary assay concepts also require some kind of fluid actuation, namely to guide the to-be-tested fluid sample into the cartridge. From the perspective of fluid handling, the simplest solution for a magnetically-controlled assay is a cartridge in which the initial transport of sample into the cartridge is effectuated by passive capillary forces. In the future, we expect that stationary-fluidic concepts will continue to gain attention, as these concepts maximally exploit the functional properties of magnetic particles to facilitate lab-on-chip integration.
In the field of integrated and magnetically actuated assays, individual process steps are being studied as well as the integration of different process steps. The use of magnetic actuation processes for integration purposes is proceeding steadily, as shown in Table 1. There are still several white spaces where actuation principles can be applied to further enhance system integration and overall analytical performance. We expect that novel actuation processes will be developed that are based on dynamic rather than static field generation. Scientifically speaking, several magnetic actuation processes have been qualitatively demonstrated but are not yet well characterized and modeled. Also, we foresee that magnetic actuation principles will be carefully attuned to specific biomaterials and reagents, and vice versa, biomaterials will be designed specifically for use in actuated assays. Concerning the magnetic particles, we foresee that particle-based assays will benefit from the ongoing optimization of particles regarding their surface bio-functionalization, surface smoothness, and their size and magnetization uniformity.136
Importantly, the performance of novel lab-on-chip analytical systems should be demonstrated by reporting dose–response curves on real-life samples. Blood is the most important matrix for in vitro diagnostic testing, yet it is a challenging fluid to work with due to the high concentrations of cells and proteins, which may cause clogging, steric hindrance, molecular interferences, non-specific adhesion, etc. Novel technologies are always first studied with spiked buffers or with diluted plasma or serum; yet the step to whole plasma, whole blood and other realistic bodily fluids should be made as quickly as possible. Furthermore, dose–response curves should be reported with sufficient statistics, so that reliable LoDs and LoQs can be determined including confidence intervals.
Overall, we see many avenues for further innovation of microfluidic Point-of-Care Testing based on magnetic particles. Magnetic particles are fundamentally suited for developing miniaturized biosensing systems and allow a range of unique stationary-fluidic system concepts. We expect that integrated magnetic actuation-based biosensing systems will have a large impact on society in the future. Such systems will allow quantitative decentralized in vitro diagnostic testing in a rapid manner with a user-friendly “sample-in result-out” type of performance, in desktop-sized and hand-held instruments. By virtue of these properties, magnetic actuation-based biosensing systems can help to improve diagnostic workflows, patient monitoring and disease management, with impact on the quality, accessibility and cost-effectiveness of future healthcare.
Acknowledgements
We thank Yang Gao, Matthias Irmscher and Toon Evers for their valuable comments. Part of this work was funded by the Dutch Technology Foundation (STW) under grant number 10458, and by NanoNextNL, a micro and nanotechnology consortium of the Government of the Netherlands and 130 partners.Notes and references
- C. M. Christensen, J. H. Grossman and J. Hwang, The Innovator's Prescription: A Disruptive Solution for Health Care, McGraw-Hill, 2009 Search PubMed.
- R. W. Forsman, Clin. Chem., 1996, 42, 813–816 CAS.
- The Value of Diagnostics Innovation, Adoption and Diffusion into Health Care, The Lewin Group, Inc, 2005, www.lewin.com/publications/publication/237 Search PubMed.
- P. von Lode, Clin. Biochem., 2005, 38, 591–606 CrossRef CAS PubMed.
- M. L. Kovarik, P. C. Gach, D. M. Ornoff, Y. L. Wang, J. Balowski, L. Farrag and N. L. Allbritton, Anal. Chem., 2012, 84, 516–540 CrossRef CAS PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- D. Wild, The Immunoassay Handbook, Elsevier, Amsterdam, 2005 Search PubMed.
- The Nucleic Acid Protocols Handbook, ed. R. Rapley, Humana Press, Totowa, New Jersey, 2000 Search PubMed.
- J. M. Nam, C. S. Thaxton and C. A. Mirkin, Science, 2003, 301, 1884–1886 CrossRef CAS PubMed.
- Z. Chen, S. M. Tabakman, A. P. Goodwin, M. G. Kattah, D. Daranciang, X. Wang, G. Zhang, X. Li, Z. Liu, P. J. Utz, K. Jiang, S. Fan and H. Dai, Nat. Biotechnol., 2008, 26, 1285–1292 CrossRef CAS PubMed.
- S. J. Osterfeld, H. Yu, R. S. Gaster, S. Caramuta, L. Xu, S. J. Han, D. A. Hall, R. J. Wilson, S. Sun, R. L. White, R. W. Davis, N. Pourmand and S. X. Wang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20637–20640 CrossRef CAS PubMed.
- G. F. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, Nat. Biotechnol., 2005, 23, 1294–1301 CrossRef CAS PubMed.
- J. Todd, B. Freese, A. Lu, D. Held, J. Morey, R. Livingston and P. Goix, Clin. Chem., 2007, 53, 1990–1995 CAS.
- R. Fan, O. Vermesh, A. Srivastava, B. K. Yen, L. Qin, H. Ahmad, G. A. Kwong, C. C. Liu, J. Gould, L. Hood and J. R. Heath, Nat. Biotechnol., 2008, 26, 1373–1378 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2012, 12, 2118–2134 RSC.
- T. G. Kang, M. A. Hulsen, P. D. Anderson, J. M. J. den Toonder and H. E. Meijer, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76, 066303 CrossRef.
- J. L. Bock, Am. J. Clin. Pathol., 2000, 113, 628–646 CrossRef CAS PubMed.
- M. A. M. Gijs, F. Lacharme and U. Lehmann, Chem. Rev., 2010, 110, 1518–1563 CrossRef CAS PubMed.
- D. Horak, M. Babic, H. Mackova and M. J. Benes, J. Sep. Sci., 2007, 30, 1751–1772 CrossRef CAS PubMed.
- C. P. Bean and J. D. Livingston, J. Appl. Phys., 1959, 30, S120 CrossRef PubMed.
- N. Pamme, Lab Chip, 2006, 6, 24–38 RSC.
- J. Pipper, Y. Zhang, P. Neuzil and T. M. Hsieh, Angew. Chem., Int. Ed., 2008, 47, 3900–3904 CrossRef CAS PubMed.
- U. Lehmann, C. Vandevyver, V. K. Parashar and M. A. M. Gijs, Angew. Chem., Int. Ed., 2006, 45, 3062–3067 CrossRef CAS PubMed.
- D. M. Bruls, T. H. Evers, J. A. H. Kahlman, P. J. W. van Lankvelt, M. Ovsyanko, E. G. M. Pelssers, J. J. H. B. Schleipen, F. K. de Theije, C. A. Verschuren, T. van der Wijk, J. B. A. van Zon, W. U. Dittmer, A. H. J. Immink, J. H. Nieuwenhuis and M. W. J. Prins, Lab Chip, 2009, 9, 3504–3510 RSC.
- M. A. Hayes, N. A. Polson and A. A. Garcia, Langmuir, 2001, 17, 2866–2871 CrossRef CAS.
- A. Rida and M. A. M. Gijs, Appl. Phys. Lett., 2004, 85, 4986–4988 CrossRef CAS PubMed.
- A. Rida and M. A. M. Gijs, Anal. Chem., 2004, 76, 6239–6246 CrossRef CAS PubMed.
- Y. Wang, J. Zhe, B. T. F. Chung and P. Dutta, Microfluid. Nanofluid., 2008, 4, 375–389 CrossRef.
- S. H. Lee, D. van Noort, J. Y. Lee, B. T. Zhang and T. H. Park, Lab Chip, 2009, 9, 479–482 RSC.
- H. Suzuki, C.-M. Ho and N. Kasagi, J. Microlithogr., Microfabr., Microsyst., 2004, 13, 779–790 Search PubMed.
- F. Lacharme, C. Vandevyver and M. A. M. Gijs, Anal. Chem., 2008, 80, 2905–2910 CrossRef CAS PubMed.
- T. Lund-Olesen, B. B. Buus, J. G. Howalt and M. F. Hansen, J. Appl. Phys., 2008, 103, 07E902 CrossRef PubMed.
- F. Wittbracht, A. Weddemann, B. Eickenberg, M. Zahn and A. Hutten, Appl. Phys. Lett., 2012, 100, 123507 CrossRef PubMed.
- Y. Moser, T. Lehnert and M. A. M. Gijs, Appl. Phys. Lett., 2009, 94, 022505 CrossRef PubMed.
- A. K. Vuppu, A. A. Garcia and M. A. Hayes, Langmuir, 2003, 19, 8646–8653 CrossRef CAS.
- A. K. Vuppu, A. A. Garcia, M. A. Hayes, K. Booksh, P. E. Phelan, R. Calhoun and S. K. Saha, J. Appl. Phys., 2004, 96, 6831–6838 CrossRef CAS PubMed.
- A. K. Vuppu, A. A. Garcia, S. K. Saha, P. E. Phelan, M. A. Hayes and R. Calhoun, Lab Chip, 2004, 4, 201–208 RSC.
- R. Calhoun, A. Yadav, P. Phelan, A. Vuppu, A. Garcia and M. Hayes, Lab Chip, 2006, 6, 247–257 RSC.
- S. Krishnamurthy, A. Yadav, P. E. Phelan, R. Calhoun, A. K. Vuppu, A. A. Garcia and M. A. Hayes, Microfluid. Nanofluid., 2008, 5, 33–41 CrossRef CAS.
- Y. Gao, M. A. Hulsen, T. G. Kang and J. M. J. den Toonder, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 86, 041503 CrossRef CAS.
- J. E. Martin, L. Shea-Rohwer and K. J. Solis, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 80, 016312 CrossRef.
- T. Roy, A. Sinha, S. Chakraborty, R. Ganguly and I. K. Puri, Phys. Fluids, 2009, 21, 027101 CrossRef PubMed.
- S. L. Biswal and A. P. Gast, Anal. Chem., 2004, 76, 6448–6455 CrossRef CAS PubMed.
- J. M. Ottino, The Kinematics of Mixing: Stretching, Chaos, and Transport, Cambridge University Press, Cambridge, England, 1989 Search PubMed.
- D. Holzinger, D. Lengemann, F. Gollner, D. Engel and A. Ehresmann, Appl. Phys. Lett., 2012, 100, 153504 CrossRef PubMed.
- Y. Gao, A. van Reenen, M. A. Hulsen, A. M. de Jong, M. W. J. Prins and J. M. J. den Toonder, Microfluid. Nanofluid., 2014, 16, 265–274 CrossRef.
- J. T. Lee, A. Abid, K. H. Cheung, L. Sudheendra and I. M. Kennedy, Microfluid. Nanofluid., 2012, 13, 461–468 CrossRef CAS PubMed.
- C. E. Smith and C. K. York, US patent, 6 027 945, 2000 Search PubMed.
- R. C. den Dulk, K. A. Schmidt, G. Sabatte, S. Liebana and M. W. J. Prins, Lab Chip, 2013, 13, 106–118 RSC.
- C. H. Chiou, D. J. Shin, Y. Zhang and T. H. Wang, Biosens. Bioelectron., 2013, 50, 91–99 CrossRef CAS PubMed.
- S. Berensmeier, Appl. Microbiol. Biotechnol., 2006, 73, 495–504 CrossRef CAS PubMed.
- G. Schreiber, G. Haran and H. X. Zhou, Chem. Rev., 2009, 109, 839–860 CrossRef CAS PubMed.
- H. X. Zhou, Q. Rev. Biophys., 2010, 43, 219–293 CrossRef PubMed.
- R. R. Gabdoulline and R. C. Wade, Curr. Opin. Struct. Biol., 2002, 12, 204–213 CrossRef CAS.
- A. Singhal, C. A. Haynes and C. L. Hansen, Anal. Chem., 2010, 82, 8671–8679 CrossRef CAS PubMed.
- A. van Reenen, A. M. de Jong and M. W. J. Prins, J. Phys. Chem. B, 2013, 117, 1210–1218 CrossRef CAS PubMed.
- K. S. Schmitz and J. M. Schurr, J. Phys. Chem., 1972, 76, 534–545 CrossRef CAS.
- M. Stenberg and H. Nygren, J. Immunol. Methods, 1988, 113, 3–15 CrossRef CAS.
- T. M. Squires, R. J. Messinger and S. R. Manalis, Nat. Biotechnol., 2008, 26, 417–426 CrossRef CAS PubMed.
- A. van Reenen, Y. Gao, A. M. de Jong, M. A. Hulsen, J. M. J. den Toonder and M. W. J. Prins, 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Okinawa, Japan, 2012, pp. 926–928 Search PubMed.
- Y. Moser, T. Lehnert and M. A. M. Gijs, Lab Chip, 2009, 9, 3261–3267 RSC.
- S. Sjolander and C. Urbaniczky, Anal. Chem., 1991, 63, 2338–2345 CrossRef CAS.
- M. A. Hayes, T. N. Polson, A. N. Phayre and A. A. Garcia, Anal. Chem., 2001, 73, 5896–5902 CrossRef CAS.
- J. W. Choi, K. W. Oh, J. H. Thomas, W. R. Heineman, H. B. Halsall, J. H. Nevin, A. J. Helmicki, H. T. Henderson and C. H. Ahn, Lab Chip, 2002, 2, 27–30 RSC.
- M. Slovakova, N. Minc, Z. Bilkova, C. Smadja, W. Faigle, C. Futterer, M. Taverna and J. L. Viovy, Lab Chip, 2005, 5, 935–942 RSC.
- B. Teste, F. Malloggi, J. M. Siaugue, A. Varenne, F. Kanoufi and S. Descroix, Lab Chip, 2011, 11, 4207–4213 RSC.
- S. Bronzeau and N. Pamme, Anal. Chim. Acta, 2008, 609, 105–112 CrossRef CAS PubMed.
- S. A. Peyman, A. Iles and N. Pamme, Lab Chip, 2009, 9, 3110–3117 RSC.
- N. Pamme, Curr. Opin. Chem. Biol., 2012, 16, 436–443 CrossRef CAS PubMed.
- K. S. Kim and J. K. Park, Lab Chip, 2005, 5, 657–664 RSC.
- H. Lee, J. Jung, S. I. Han and K. H. Han, Lab Chip, 2010, 10, 2764–2770 RSC.
- R. Ganguly, T. Hahn and S. Hardt, Microfluid. Nanofluid., 2010, 8, 739–753 CrossRef CAS.
- K. Tanaka and H. Imagawa, Talanta, 2005, 68, 437–441 CrossRef CAS PubMed.
- J. T. Lee, L. Sudheendra and I. M. Kennedy, Anal. Chem., 2012, 84, 8317–8322 CrossRef CAS PubMed.
- H. C. Tekin and M. A. M. Gijs, Lab Chip, 2013, 13, 4711–4739 RSC.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29 Suppl 1, S49–52 Search PubMed.
- S. Chandran and R. S. Singh, Pharmazie, 2007, 62, 4–14 CAS.
- D. M. Rissin, C. W. Kan, T. G. Campbell, S. C. Howes, D. R. Fournier, L. Song, T. Piech, P. P. Patel, L. Chang, A. J. Rivnak, E. P. Ferrell, J. D. Randall, G. K. Provuncher, D. R. Walt and D. C. Duffy, Nat. Biotechnol., 2010, 28, 595–599 CrossRef CAS PubMed.
- A. D. Wellman and M. J. Sepaniak, Anal. Chem., 2007, 79, 6622–6628 CrossRef CAS PubMed.
- M. Herrmann, T. Veres and M. Tabrizian, Anal. Chem., 2008, 80, 5160–5167 CrossRef CAS PubMed.
- R. S. Sista, A. E. Eckhardt, V. Srinivasan, M. G. Pollack, S. Palanki and V. K. Pamula, Lab Chip, 2008, 8, 2188–2196 RSC.
- A. H. Ng, K. Choi, R. P. Luoma, J. M. Robinson and A. R. Wheeler, Anal. Chem., 2012, 84, 8805–8812 CrossRef CAS PubMed.
- C. W. Kan, A. J. Rivnak, T. G. Campbell, T. Piech, D. M. Rissin, M. Mosl, A. Peterca, H. P. Niederberger, K. A. Minnehan, P. P. Patel, E. P. Ferrell, R. E. Meyer, L. Chang, D. H. Wilson, D. R. Fournier and D. C. Duffy, Lab Chip, 2012, 12, 977–985 RSC.
- Z. Fu, G. Shao, J. Wang, D. Lu, W. Wang and Y. Lin, Anal. Chem., 2011, 83, 2685–2690 CrossRef CAS PubMed.
- D. Tang, R. Yuan and Y. Chai, Clin. Chem., 2007, 53, 1323–1329 CAS.
- J. Do and C. H. Ahn, Lab Chip, 2008, 8, 542–549 RSC.
- D. Witters, K. Knez, F. Ceyssens, R. Puers and J. Lammertyn, Lab Chip, 2013, 13, 2047–2054 RSC.
- D. M. Rissin, C. W. Kan, L. Song, A. J. Rivnak, M. W. Fishburn, Q. Shao, T. Piech, E. P. Ferrell, R. E. Meyer, T. G. Campbell, D. R. Fournier and D. C. Duffy, Lab Chip, 2013, 13, 2902–2911 RSC.
- Y. K. Hahn and J. K. Park, Lab Chip, 2011, 11, 2045–2048 RSC.
- R. Gottheil, N. Baur, H. Becker, G. Link, D. Maier, N. Schneiderhan-Marra and M. Stelzle, Biomed. Microdevices, 2013, 16, 163–172 CrossRef PubMed.
- S. M. Berry, E. T. Alarid and D. J. Beebe, Lab Chip, 2011, 11, 1747–1753 RSC.
- K. Sur, S. M. McFall, E. T. Yeh, S. R. Jangam, M. A. Hayden, S. D. Stroupe and D. M. Kelso, J. Mol. Diagn., 2010, 12, 620–628 CrossRef CAS PubMed.
- Y. Zhang, S. Park, K. Liu, J. Tsuan, S. Yang and T. H. Wang, Lab Chip, 2011, 11, 398–406 RSC.
- O. Strohmeier, A. Emperle, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 146–155 RSC.
- R. Sista, Z. Hua, P. Thwar, A. Sudarsan, V. Srinivasan, A. Eckhardt, M. Pollack and V. Pamula, Lab Chip, 2008, 8, 2091–2104 RSC.
- M. Shikida, K. Takayanagi, H. Honda, H. Ito and K. Sato, J. Micromech. Microeng., 2006, 16, 1875–1883 CrossRef.
- H. Bordelon, N. M. Adams, A. S. Klemm, P. K. Russ, J. V. Williams, H. K. Talbot, D. W. Wright and F. R. Haselton, ACS Appl. Mater. Interfaces, 2011, 3, 2161–2168 CAS.
- B. P. Casavant, D. J. Guckenberger, S. M. Berry, J. T. Tokar, J. M. Lang and D. J. Beebe, Lab Chip, 2013, 13, 391–396 RSC.
- R. C. den Dulk, K. A. Schmidt, R. Gill, J. C. B. Jongen and M. W. J. Prins, Proc. MicroTAS 2010 Int. Conf., 2010, pp. 665–667 Search PubMed.
- D. J. Shin, L. Chen and T. H. Wang, Proc. MicroTAS 2013 Int. Conf., 2013, pp. 1350–1352 Search PubMed.
- H. Tsuchiya, M. Okochi, N. Nagao, M. Shikida and H. Honda, Sens. Actuators, B, 2008, 130, 583–588 CrossRef CAS PubMed.
- G. Degre, E. Brunet, A. Dodge and P. Tabeling, Lab Chip, 2005, 5, 691–694 RSC.
- A. Ranzoni, G. Sabatte, L. J. van IJzendoorn and M. W. J. Prins, ACS Nano, 2012, 6, 3134–3141 CrossRef CAS PubMed.
- J. Baudry, C. Rouzeau, C. Goubault, C. Robic, L. Cohen-Tannoudji, A. Koenig, E. Bertrand and J. Bibette, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16076–16078 CrossRef CAS PubMed.
- S. Y. Park, H. Handa and A. Sandhu, J. Appl. Phys., 2009, 105, 07B526 Search PubMed.
- S. Y. Park, H. Handa and A. Sandhu, Nano Lett., 2010, 10, 446–451 CrossRef CAS PubMed.
- S. Y. Park, P. J. Ko, H. Handa and A. Sandhu, J. Appl. Phys., 2010, 107, 09B324 Search PubMed.
- D. Issadore, C. Min, M. Liong, J. Chung, R. Weissleder and H. Lee, Lab Chip, 2011, 11, 2282–2287 RSC.
- I. Koh, R. Hong, R. Weissleder and L. Josephson, Angew. Chem., Int. Ed., 2008, 47, 4119–4121 CrossRef CAS PubMed.
- A. Ranzoni, J. J. H. B. Schleipen, L. J. van IJzendoorn and M. W. J. Prins, Nano Lett., 2011, 11, 2017–2022 CrossRef CAS PubMed.
- Y. Gao, A. van Reenen, M. A. Hulsen, A. M. de Jong, M. W. J. Prins and J. M. J. den Toonder, Lab Chip, 2013, 13, 1394–1401 RSC.
- W. U. Dittmer, P. de Kievit, M. W. J. Prins, J. L. Vissers, M. E. Mersch and M. F. Martens, J. Immunol. Methods, 2008, 338, 40–46 CrossRef CAS PubMed.
- V. N. Morozov and T. Y. Morozova, Anal. Chim. Acta, 2006, 564, 40–52 CrossRef CAS PubMed.
- R. De Palma, C. Liu, F. Barbagini, G. Reekmans, K. Bonroy, W. Laureyn, G. Borghs and G. Maes, J. Phys. Chem. C, 2007, 111, 12227–12235 CAS.
- R. De Palma, G. Reekmans, W. Laureyn, G. Borghs and G. Maes, Anal. Chem., 2007, 79, 7540–7548 CrossRef CAS PubMed.
- V. N. Morozov, S. Groves, M. J. Turell and C. Bailey, J. Am. Chem. Soc., 2007, 129, 12628–12629 CrossRef CAS PubMed.
- Y. M. Shlyapnikov, E. A. Shlyapnikova, M. A. Simonova, A. O. Shepelyakovskaya, F. A. Brovko, R. L. Komaleva, E. V. Grishin and V. N. Morozov, Anal. Chem., 2012, 84, 5596–5603 CrossRef CAS PubMed.
- T. Y. Morozova and V. N. Morozov, Anal. Biochem., 2008, 374, 263–271 CrossRef CAS PubMed.
- H. C. Tekin, M. Cornaglia and M. A. M. Gijs, Lab Chip, 2013, 13, 1053–1059 RSC.
- C. B. Kriz, K. Radevik and D. Kriz, Anal. Chem., 1996, 68, 1966–1970 CrossRef CAS PubMed.
- Y. R. Chemla, H. L. Grossman, Y. Poon, R. McDermott, R. Stevens, M. D. Alper and J. Clarke, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14268–14272 CrossRef CAS PubMed.
- D. R. Baselt, G. U. Lee, M. Natesan, S. W. Metzger, P. E. Sheehan and R. J. Colton, Biosens. Bioelectron., 1998, 13, 731–739 CrossRef CAS.
- M. Koets, T. van der Wijk, J. T. W. M. van Eemeren, A. van Amerongen and M. W. J. Prins, Biosens. Bioelectron., 2009, 24, 1893–1898 CrossRef CAS PubMed.
- R. S. Gaster, D. A. Hall and S. X. Wang, Lab Chip, 2011, 11, 950–956 RSC.
- T. Aytur, J. Foley, M. Anwar, B. Boser, E. Harris and P. R. Beatty, J. Immunol. Methods, 2006, 314, 21–29 CrossRef CAS PubMed.
- F. Colle, D. Vercruysse, S. Peeters, C. Liu, T. Stakenborg, L. Lagae and J. Del-Favero, Lab Chip, 2013, 13, 4257–4262 RSC.
- Y. Wang, J. Dostalek and W. Knoll, Anal. Chem., 2011, 83, 6202–6207 CrossRef CAS PubMed.
- D. Leckband and J. Israelachvili, Q. Rev. Biophys., 2001, 34, 105–267 CrossRef CAS.
- C. Danilowicz, D. Greenfield and M. Prentiss, Anal. Chem., 2005, 77, 3023–3028 CrossRef CAS PubMed.
- A. Jacob, L. J. van IJzendoorn, A. M. de Jong and M. W. J. Prins, Anal. Chem., 2012, 84, 9287–9294 CAS.
- I. D. Vilfan, J. Lipfert, D. A. Koster, S. G. Lemay and N. H. Dekker, Magnetic Tweezers for Single-Molecule Experiments: Springer Handbook of Single-Molecule Biophysics, Springer, New York, 2009 Search PubMed.
- I. De Vlaminck and C. Dekker, Annu. Rev. Biophys., 2012, 41, 453–472 CrossRef CAS PubMed.
- A. van Reenen, F. Gutierrez-Mejia, L. J. van IJzendoorn and M. W. J. Prins, Biophys. J., 2013, 104, 1073–1080 CrossRef CAS PubMed.
- X. J. A. Janssen, A. J. Schellekens, K. van Ommering, L. J. van Ijzendoorn and M. W. J. Prins, Biosens. Bioelectron., 2009, 24, 1937–1941 CrossRef CAS PubMed.
- A. van Reenen, Y. Gao, A. H. Bos, A. M. de Jong, M. A. Hulsen, J. M. J. den Toonder and M. W. J. Prins, Appl. Phys. Lett., 2013, 103, 043704 CrossRef PubMed.
- R. C. Ebersole, US Pat. 4219335, 1978 Search PubMed.
- G. U. Lee, S. Metzger, M. Natesan, C. Yanavich and Y. F. Dufrene, Anal. Biochem., 2000, 287, 261–271 CrossRef CAS PubMed.
- D. A. Boas, C. Pitris and N. Ramanujam, Handbook of Biomedical Optics, CRC Press, Boca Raton, FL, 2011 Search PubMed.
Footnotes |
| † In the scientific literature, the terms ‘magnetic particle’ and ‘magnetic bead’ are often interchangeably used. We use ‘magnetic particle’ because it is more general, as ‘magnetic bead’ mostly relates to spherical particles made of composite material. |
| ‡ In the scientific literature, the terms ‘particle agglutination’, ‘particle aggregation’ and ‘particle clustering’ are interchangeably used. |
| This journal is © The Royal Society of Chemistry 2014 |