 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Light-assisted direct-write of 3D functional biomaterials
Kolin C.
Hribar
,
Pranav
Soman
,
John
Warner
,
Peter
Chung
and
Shaochen
Chen
*
Department of NanoEngineering, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA. E-mail: chen168@eng.ucsd.edu
First published on 20th November 2013
Abstract
Light-assisted 3D direct-printing of biomaterials and cellular-scaffolds has the potential to develop novel lab-on-a-chip devices (LOCs) for a variety of biomedical applications, from drug discovery and diagnostic testing to in vitro tissue engineering and regeneration. Direct-writing describes a broad family of fabrication methods that typically employ computer-controlled translational stages to manufacture structures at multi-length scales. This review focuses on light-assisted direct-write fabrication for generating 3D functional scaffolds with precise micro- and nano-architecture, using both synthetic as well as naturally derived biomaterials. Two bioprinting approaches are discussed in detail – projection printing and laser-based systems – where each method is capable of modulating multiple scaffold parameters, such as 3D architecture, mechanical properties (e.g. stiffness), Poisson's ratio, chemical gradients, biological cell distributions, and porosity. The light-assisted direct-writing techniques described in this review provide the reader with alternative approaches to fabricate 3D biomaterials for utility in LOCs.
I. Introduction
Over the years, lab-on-a-chip platforms (LOCs) have been extensively developed for the measurement of fundamental biological components (blood glucose, coagulation, cardiac), health biomarkers (metabolic disorders), and infectious agents (pathogens, viruses, anthrax), and have been used for in vitro diffusion models (e.g. protein kinetic measurements, glucose concentration monitoring) as well as human tissue models (heart, lung, liver on a chip).1,2 More recently, researchers have sought to develop LOCs with 3D multicellular environments better mimicking native tissue while permitting user controlled modification.2,3 Bioprinting platforms for these types of tissue engineered constructs have immense potential in generating more physiologically relevant LOCs, enabling the rapid screening of toxins, drugs, and ligands in addition to investigating fundamental cell biology in a more native, yet controllable micro/nano 3D environment.4,5The evolving fields of tissue engineering and regeneration, and in larger context, bioengineering and its associated areas, have advanced in tissue culture in order to mimic one or several aspects of the physiological/pathophysiological environment.6–9 This refinement has progressed from two-dimensional (2D) extracellular matrix (ECM)-coated substrates to three-dimensional (3D) patterned scaffolds for cell seeding and encapsulation. 2D refers to flat substrates with cell–substrate interactions primarily in the XY plane and negligible interactions along the z-direction. 3D substrates consist of either defined or random anisotropic architecture along the z-direction. Several studies have demonstrated that cells grown in 2D cultures display inconsistencies with the physiological in vivo environment with respect to morphology, cell–cell and cell–ECM contacts, proliferation and differentiation.10 Thus, research has prompted the development of various biofabrication methods in order to more effectively recreate the native 3D tissue.11
3D biofabrication is primarily divided into two types: computer-assisted and process-directed techniques. Process-driven fabrication methods – those which do not utilize computer control or direct placement – include freeze-drying,12 salt leaching,13 electrospinning,14 porogen melting15 gas foaming,16 and fiber deposition.17 These methods allow control over bulk physical properties (e.g. material stiffness, swelling, etc.), however the resulting internal architecture is relatively uncontrolled. Computer-assisted direct-writing approaches, on the other hand, are capable of precisely dictating the internal architecture at the micro- and nanoscale, and thus facilitating greater control at the cellular and subcellular level.
Direct-writing techniques, typically referred to as free-form fabrication or rapid prototyping, are well suited to manufacture 3D scaffolds with orthogonal control over fabrication parameters.18,19 In most cases, a user-defined 3D structure is modeled using a computer software and broken down into a series of transverse-plane image slices. These slices are created as thin 2D layers for stacking in a layer-by-layer fashion (additive manufacturing), and a 3D scaffold is fabricated according to these image slices by translating either the computer-controlled stage or the deposition source in XYZ directions. Direct-writing allows the investigation of one or several biophysical parameters such as internal-architecture and mechanical properties. This approach also permits the use of a wide array of physiological components, such as biochemicals and encapsulated cells, in a modular fashion. Within the confines of direct-writing, the utility of light (e.g. laser or UV) offers a precise, rapid, and cost-effective way to fabricate 3D bio-structures at the micro- and nanoscale. This review will discuss two light-assisted direct-write bioprinting platforms and applications of each: (A) projection printing and (B) laser-based systems, in addition to highlighting some commonly used biomaterials.
Biomaterials
Numerous monomers and a selection of photoinitiators provide many permutations of materials for light-assisted fabrication systems.19 Monomers of different chain lengths and chemical species at varying concentrations can be used to tune mechanical properties, porosity, and osmotic swelling of the resulting polymerized gels. Modulation of light intensity can also vary the degree of polymerization, additionally affecting these parameters. Though many conventional hydrogels such as agarose and polyacrylamide have been used in light-assisted printing, we will focus on three extensively-utilized, biocompatible, photocurable biomaterials which form hydrogels through free-radical photopolymerization: synthetic poly(ethylene glycol) diacrylate (PEGDA),20,21 and naturally-derived gelatin methacrylate (GelMA)22 and hyaluronic acid (HA).23 For a more complete list of polymers used in hydrogel fabrication, the reader is asked to consult the ref. 24 and 25.PEGDA provides an excellent material for biomedical applications because of its high water content, biocompatibility and tunable mechanical properties.26 Like many monomers, PEGDA may be synthesized at different molecular weights (typically 700–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da, in accordance to the number of polymer chains) and poly-distributions to generate polymerized gels of varying crosslinking densities and materials properties (e.g. stiffness, swelling, porosity). Moreover, multiple PEGDA monomers may be mixed at different concentrations to further alter the aforementioned material properties (e.g. 10 kDa PEGDA mixed with 700 Da PEGDA). Some synthetic materials like PEGDA are nondegradable, however chemical modification or mixing with other materials (e.g. DTT) may achieve a predictable degradation effect.27 For cell seeding, PEG scaffolds require the grafting of adhesive peptide sequences (e.g. RGDS and YIGSR) or adhesive proteins (e.g. fibronectin and laminin).
000 Da, in accordance to the number of polymer chains) and poly-distributions to generate polymerized gels of varying crosslinking densities and materials properties (e.g. stiffness, swelling, porosity). Moreover, multiple PEGDA monomers may be mixed at different concentrations to further alter the aforementioned material properties (e.g. 10 kDa PEGDA mixed with 700 Da PEGDA). Some synthetic materials like PEGDA are nondegradable, however chemical modification or mixing with other materials (e.g. DTT) may achieve a predictable degradation effect.27 For cell seeding, PEG scaffolds require the grafting of adhesive peptide sequences (e.g. RGDS and YIGSR) or adhesive proteins (e.g. fibronectin and laminin).
Naturally derived hydrogels made from gelatin methacrylate (GelMA) have biologically active sequences that bind key integrins, enabling cells to adhere and migrate onto the structure.22,28 GelMA is generally a xenogenic modified macromer, therefore relatively inexpensive depending on the source and has the associated limitations in vivo. Hyaluronic acid (HA), a naturally occurring native ECM component and FDA-approved biomaterial, is a non-immunogenic polymer known to be important in wound healing. For instance, exogenous HA has been shown to reduce scar formation and promote regeneration in peripheral nerve injuries.23 Since various biochemical moieties can be functionally introduced along the HA polymer backbone, a photopolymerizable form of HA has been created through the addition of methacrylate groups, termed glycidyl methacrylate-modified HA (GMHA). GMHA scaffold surfaces can be further modified to incorporate the cell-adhesive protein laminin. The library of photopolymerizable materials continues to expand with the development and modification of new and existing monomers/macromers, respectively, such as the aforementioned GMHA and GelMA, and thus light-assisted printing has the ability to work with an abundance of materials.
II. Projection printing systems
Digital mask (i.e. “maskless”) projection printing is a type of stereolithography which uses a digital micro-mirror device (DMD) found in conventional computer projectors to polymerize and solidify a photosensitive liquid prepolymer using either UV or other light sources.19,21,29–36Fig. 1A shows a version of the maskless projection printing system called the dynamic optical projection stereolithography (DOPsL) platform.20,21,37–42 The “maskless” or digital mask option allows for the use of controllable and interchangeable reflected light patterns rather than static, more expensive physical masks (such as those used in photolithography). The system spatially modulates collimated UV light using a DMD chip (1920 × 1080 resolution) to project custom-defined optical patterns onto a photocurable prepolymer solution. The DMD chip serves as an array of reflective coated aluminum micro-mirrors, capable of redirecting light to two states [0,1], tilted with two bias electrodes to form angles of either +12° or −12° with respect to the surface. Illumination from the light source reflects into the projection lens only when the micro-mirror is in its arbitrary “on” state. In the “off” state, the pixel appears dark as the illuminated light is reflected away from the projection lens.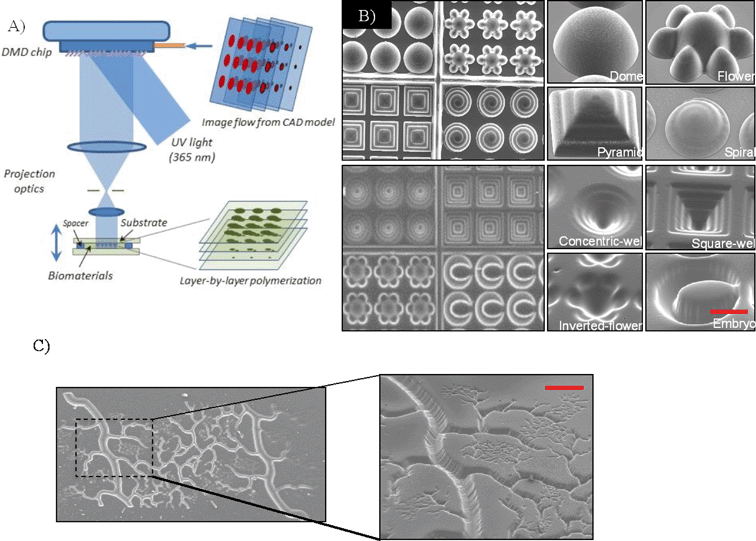 | ||
| Fig. 1 (A) Schematic of a projection printing setup called dynamic optical projection stereolithography (DOPsL): UV-light illuminates the DMD mirror system, which generates an optical pattern according to the image flow from the control computer. The optical pattern is projected through an optical lens and onto the photosensitive biomaterial to fabricate a 3D scaffold in a layer-by-layer manner. (B) SEM images of the concave and convex features (e.g. domes, microwells, etc.) using PEGDA demonstrating the versatility of the biofabrication system.41 (C) SEM image of a complex vasculature in PEGDA fabricated from a CAD file. Scale bar = 100 μm. | ||
To generate 3D structures, projection stereolithography platforms such as DOPsL employ a layer-by-layer fabrication procedure. Often, a 3D computer rendering (made with a CAD software or CT scans) is deconstructed into a series of evenly spaced planes, or layers. The pattern for each layer is displayed on the DMD chip, exposing UV light onto the photocurable material. After one layer is patterned, the substrate is lowered using an automated stage and the next pattern is displayed. The user has control over the stage speed, intensity of the light, and height of the structure, allowing for the fabrication of various complex structures, such as spiral domes, pyramids and microwells (Fig. 1B).41Fig. 1C demonstrates a complex vascular structure fabricated in PEGDA using this technology, another example of its versatility. As previously mentioned, mimicking the native tissue microenvironment with respect to architecture and material properties is key to the design of these 3D biostructures.
We will illustrate projection printing's increasing importance and utility in the tissue engineering field with several examples. From Gauvin et al., hydrogels comprised of GelMA were fabricated in 3D log-pile and hexagonal layered constructs (Fig. 2A).22 Mechanical properties were varied by the geometry and prepolymer concentration (Fig. 2B), and following, cell migration and proliferation into the scaffolds were monitored (Fig. 2C). Log-pile and hexagonal structures displayed different moduli even when the prepolymer solution remained constant (10% GelMA). Importantly, the results from this study demonstrated the capability of the projection printing systems to generate cell-compatible scaffolds with tailored mechanical properties (by either varying prepolymer concentration or the micro-architecture). In another example, Lin et al. utilized projection printing to investigate cellular responses to varying scaffold porosities (Fig. 3A).34 After encapsulating adipose-derived stem cells within the structures and incubating for seven days, an MTS assay reported a higher cell viability and activity of the cells in porous structures than in solid, non-porous structures (Fig. 3B).
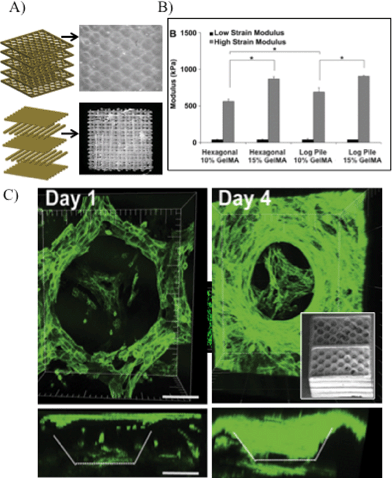 | ||
| Fig. 2 (A) Schematic shows layer-by-layer manufacturing of log-pile and hexagonal internal architecture. Optical images depict 3D scaffolds using GelMA biomaterial. (B) Mechanical properties of scaffolds having hexagonal or log-pile structures using various GelMA percentages. Prepolymer concentration and engineered structures can be used to tailor the mechanical properties of the GelMA scaffolds. * indicates statistical significance (p < 0.05). (C) 3D confocal images showing scaffold coverage by the HUVEC-GFP cells and cell penetration into the porous structure as a function of time; inset: SEM image of hexagonal layered scaffold.22 Scale = 100 μm. | ||
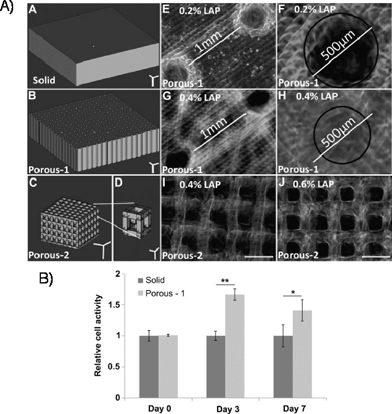 | ||
| Fig. 3 (A) 3D gels of varying porosity fabricated by projection stereolithography, with the CAD drawings and resulting internal architectures. Scale bar = 500 μm unless otherwise indicated. (B) Relative cell activity assessed by MTS assay, using either solid or porous scaffolds.34 | ||
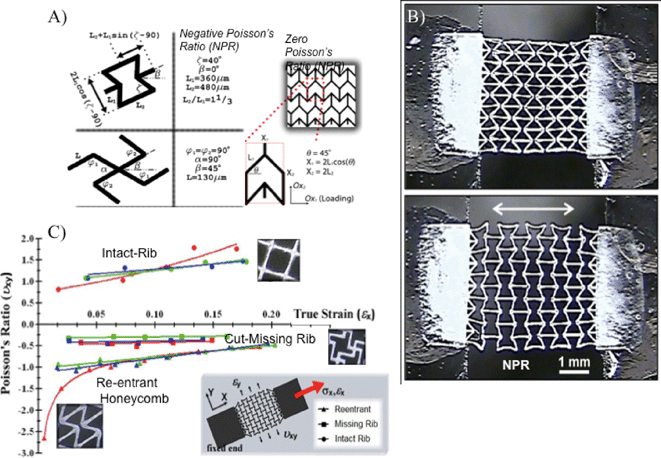 | ||
| Fig. 4 (A) Unit-cell parameters and relevant dimensional parameters of NPR (reentrant honeycomb, cut missing rib) and ZPR (semi-re-entrant) architectures. The walls of the unit-cells (denoted as ribs) are approximately 40 μm wide and 50–100 μm deep. (B) PEGDA scaffolds with NPR expand with application of strain in the X-axis (white arrow). Scale = 1 mm. (C) Measured Poisson's ratio as a function of true strain for single-layer PEGDA scaffolds composed of the reentrant, missing-rib, and intact-rib (control) unit-cell geometries.42 | ||
Another parameter in tissue engineering – Poisson's ratio (PR) – has also been investigated using projection printing.39,40,42 In general, the mechanical properties of a biomaterial scaffold can be quantitatively described by its elastic modulus and PR. Elastic modulus of the underlying substrate describes the material's elastic behavior during loading, while PR refers to the degree at which the scaffold contracts/expands in the transverse direction. Previously, research has extensively reported on the connection between a substrate's elastic modulus and cell response.43,44 PR has been less explored with conventional manufacturing approaches (e.g. annealing of polyurethane foams), because they offer little control over the microarchitecture.45,46 Light-assisted direct-writing, on the other hand, can provide precise control over this parameter. For instance, our lab used the DOPsL platform to fabricate unit-cell geometries for negative Poissson's ratio (NPR) (re-entrant structure, missing rib model) and zero Poisson ratio (ZPR) (semi-reentrant structure) scaffolds (Fig. 4A). Fig. 4B shows a structure undergoing tensile stress and expanding in the transverse direction, thereby demonstrating NPR. Analysis of the PR effect according to several unit-cells is documented in Fig. 4C. Scaffolds exhibiting NPR (those which expand upon application of tensile stress) or ZPR may be more suitable for emulating the behavior of certain native tissues (e.g. expansion in blood vessels), and could be further investigated with projection printing.
These examples and others serve to demonstrate the versatility of projection printing systems in fabricating complex 3D structures of varying topographical features, mechanical properties, and porosity. Additionally, projection printing offers superior fabrication speeds (e.g. DOPsL fabricates within seconds) as compared to serial writing of two/multiple-photon polymerization.19,41 However, limitations include the resolution feature size (due to limitations of the projection lens and material swelling), the requirement of using photopolymerizable materials, and the effect of UV exposure to cells and photosensitive biomaterials.19,47 Improved optics will continue to enhance the resolution, and the expanding library of photopolymerizable materials will allow for increased scaffold complexity.
III. Laser-based techniques
Many of the concepts introduced with the projection-based systems, including material selection, geometry optimization, cell seeding and encapsulation design, also apply to the laser-based systems.18 Historically, laser-based printing techniques such as laser-direct-writing,48,49 laser-induced forward transfer (LIFT),50 matrix-assisted pulsed laser evaporation (MAPLE)51etc., have been used to spatially pattern cells in 2D,52,53 with limitations of low cell viability and throughput. Recently, 3D laser bioprinting has evolved to additive manufacturing (e.g. layer-by-layer).54–563D laser direct-writing incorporates a CAD model numerically sliced into a series of 2D layers, and fabricates 3D structures utilizing a motorized stage (controlled in XYZ) to manipulate the sample or laser (Fig. 5A).33,57 Laser-based fabrication differs from projection printing primarily in its light source,58–60 where a laser focused through an objective lens is used to crosslink and polymerize a liquid biopolymer. Writing width can be modulated and controlled by exposure energy – dictated by the beam size and laser power – and writing speed. After the first layer is crosslinked, the stage moves downward and a new layer of polymer is solidified according to the design. One such system, termed SLA (stereolithography apparatus), employs a focused single-photon UV laser to polymerize photocurable materials.57 Chan et al. used SLA for generating controlled 3D structures to co-culture a heterogeneous cell distribution and assess long-term cell viability (Fig. 5B). In a similar laser setup, Mapili et al. demonstrated a multilayer PEG hydrogel scaffold functionalized with heparin or RGD-peptide sequences for cell adhesion (Fig. 5C).61
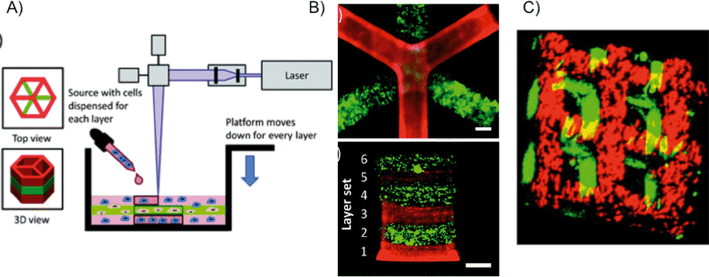 | ||
| Fig. 5 (A) Schematic of laser stereolithography using SLA (stereolithography apparatus).57 (B) Resulting SLA-fabricated 3D hydrogels with six layers.57 Scale bars = 1 mm. (C) Spatially-patterned 3D scaffolds fabricated by laser-assisted stereolithography.61 | ||
Two-photon polymerization (TPP) is another type of laser-based direct-writing which uses focused near-infrared (NIR) femtosecond laser pulses (wavelength ~800 nm) to trigger crosslinking of a photosensitive biomaterial.62 In this fabrication technique, simultaneous absorption of two NIR photons generates free radicals to initiate the polymerization of volume-blocks (voxels).63–65 A distinct advantage of this system is the nonlinear nature of the laser–material interaction, achieving feature sizes below the diffraction limit of applied light. This process allows for better resolution at the expense of slower speeds and limited scalability (due to restrictions of the objective lens' working distance) compared to single photon polymerization (e.g. SLA) (Fig. 6A). Fig. 6B shows a typical TPP laser setup. In tissue engineering applications, TPP has been exploited to generate precise microscale biomaterial structures. For instance, in Fig. 6C, TPP was used to fabricate a 3D log-pile structure (1 micron width lines) made of PEGDA with spacing of 8 μm. In another example, a gradient array of microdots (2–10 microns in diameter) was fabricated using a mixture of PEGDA and Cultrex 3-D Culture Matrix, a type of standardized basement membrane extract (Fig. 6D).66 Lee et al. also demonstrated TPP fabrication of RGD-modified hydrogels to dictate cell migration in 3D (Fig. 6E).67 These examples demonstrate TPP's ability to chemically pattern and generate biostructures at cellular and subcellular scales.
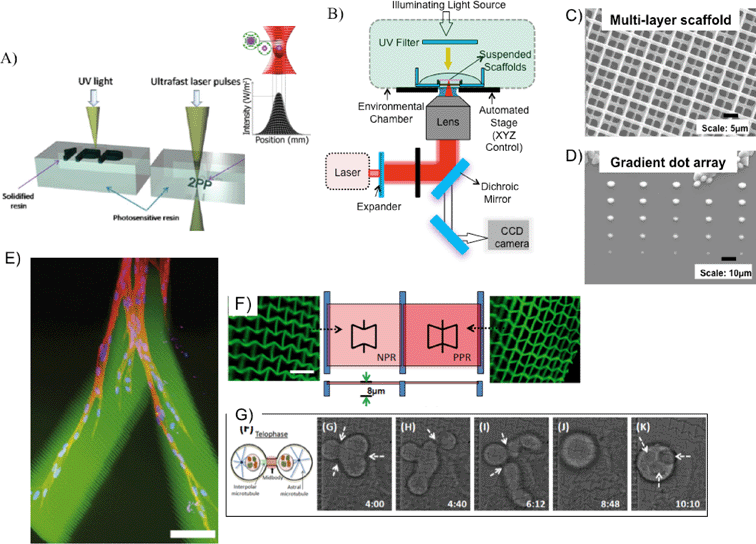 | ||
| Fig. 6 (A) Principle of operation for single- and two-photon polymerization.69 (B) Schematic of femtosecond laser fabrication set-up. (C) Multi-layer log-pile scaffold with ~1 μm features.66 (D) Gradient dot array at sub-micron resolution (the largest dot size ~4 μm, with spacing ~20 μm) fabricated by decreasing the laser-power from top to bottom. Biochemicals such as growth factors can be incorporated in these arrays.66 (E) 3D migration of cells within an RGD-modified PEG hydrogel, fabricated by TPP. Scale bar = 100 μm.67 (F) NPR and PPR structures patterned with TPP, and (G) time-resolved single cell studies on an NPR structure (for details on this study, please refer to ref. 68). | ||
Additionally, one can use TPP to explore the interaction of topographical features and single cell response. Similar to a study described in the projection printing section, Zhang et al. fabricated PEGDA-derived scaffolds exhibiting either NPR or PPR (negative or positive Poisson's ratio, respectively) (Fig. 6F).68 Cell motility, orientation, and proliferation varied between NPR and PPR constructs, suggesting that PR plays a role in cell fate (Fig. 6G); further tests are needed to promote this claim, however. Importantly, single cell studies are possible with TPP due to its high resolution of fabrication, compared to single photon or projection printing.
Conclusion
Both projection printing (e.g. DOPsL) and laser-based direct-writing systems (SLA, TPP) offer promising technologies to fabricate 3D bioconstructs with precise micro- and nano-architecture. Though these processes require photopolymerizable materials, the library of such materials continues to expand, accommodating for various scaffold properties (e.g. stiffness, porosity, swelling) in a 3D setting. Additionally, projection printing allows for the rapid printing of various complex structures, demonstrating its potential for high throughput screening. Though typically slower than projection printing (and SLA), TPP provides a high-resolution alternative to direct-writing for the fabrication of nanoscale features, and could play a significant role in single cell analysis in the years to come. More interesting yet, combining several platforms such as projection printing and TPP could enable researchers to study a multitude of parameters in 3D. Light-assisted direct-writing could also complement non-light-assisted direct-writing platforms (e.g. nozzle-based or extrusion systems) for generating precise nano/microscale features and chemical patterning within a larger scaffold. In the years ahead, we welcome the convergence of multiple printing technologies in the pursuit of growing physiologically relevant 3D tissue constructs in vitro (i.e. tissue-on-a-chip). Such in vitro biomimetic LOCs will enable us to more accurately answer basic cell biology questions and monitor various testing parameters (e.g. drug toxicity/discovery, (patho)physiology) prior to more expensive in vivo models.Acknowledgements
The project described was supported by grants (EB012597 and EB017876) from the National Institute of Biomedical Imaging and Bioengineering and grants (CMMI-1130894, CMMI-1120795) from the US National Science Foundation.References
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2012, 12, 2118–2134 RSC.
- A. M. Ghaemmaghami, M. J. Hancock, H. Harrington, H. Kaji and A. Khademhosseini, Drug Discovery Today, 2012, 17, 173–181 CrossRef CAS PubMed.
- D. E. Ingber and G. M. Whitesides, Lab Chip, 2012, 12, 2089–2090 RSC.
- M. Baker, Nature, 2011, 471, 661–665 CrossRef CAS PubMed.
- D. Huh, Y.-S. Torisawa, G. A. Hamilton, H. J. Kim and D. E. Ingber, Lab Chip, 2012, 12, 2156–2164 RSC.
- I. R.-D. Jorge, Z. Bimeng, R. Daniel, S. Zhi-dong and X. Tao, Biofabrication, 2012, 4, 035001 CrossRef PubMed.
- P. X. Ma, Adv. Drug Delivery Rev., 2008, 60, 184–198 CrossRef CAS PubMed.
- R. Langer and J. P. Vacanti, Science, 1993, 260, 920–926 CAS.
- L. G. Griffith and G. Naughton, Science, 2002, 295, 1009–1014 CrossRef CAS PubMed.
- A. J. Engler, M. A. Griffin, S. Sen, C. G. Bönnemann, H. L. Sweeney and D. E. Discher, J. Cell Biol., 2004, 166, 877–887 CrossRef CAS PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- F. J. O'Brien, B. A. Harley, I. V. Yannas and L. Gibson, Biomaterials, 2004, 25, 1077–1086 CrossRef CAS.
- S. B. Lee, Y. H. Kim, M. S. Chong, S. H. Hong and Y. M. Lee, Biomaterials, 2005, 26, 1961–1968 CrossRef CAS PubMed.
- X. Zong, H. Bien, C.-Y. Chung, L. Yin, D. Fang, B. S. Hsiao, B. Chu and E. Entcheva, Biomaterials, 2005, 26, 5330–5338 CrossRef CAS PubMed.
- A. S. P. Lin, T. H. Barrows, S. H. Cartmell and R. E. Guldberg, Biomaterials, 2003, 24, 481–489 CrossRef CAS.
- R. Nazarov, H.-J. Jin and D. L. Kaplan, Biomacromolecules, 2004, 5, 718–726 CrossRef CAS PubMed.
- L. Moroni, J. R. de Wijn and C. A. van Blitterswijk, Biomaterials, 2006, 27, 974–985 CrossRef CAS PubMed.
- I. Antoniac, T. Billiet, M. Vandenhaute, J. Schelfhout, S. Vlierberghe and P. Dubruel, in Biologically Responsive Biomaterials for Tissue Engineering, Springer, New York, 2013, pp. 201–249 Search PubMed.
- T. Billiet, M. Vandenhaute, J. Schelfhout, S. Van Vlierberghe and P. Dubruel, Biomaterials, 2012, 33, 6020–6041 CrossRef CAS PubMed.
- L. H. Han, G. Mapili, S. Chen and K. Roy, J. Manuf. Sci. E–T. ASME, 2008, 130, 021005 CrossRef.
- Y. Lu, G. Mapili, G. Suhali, S. C. Chen and K. Roy, J. Biomed. Mater. Res., Part A, 2006, 77, 396–405 CrossRef PubMed.
- R. Gauvin, Y.-C. Chen, J. W. Lee, P. Soman, P. Zorlutuna, J. Nichol, S. C. Chen and A. Khademhosseini, Biomaterials, 2012, 33, 3824–3834 CrossRef CAS PubMed.
- S. Suri, L. H. Han, W. Zhang, A. Singh, S. C. Chen and C. E. Schmidt, Biomed. Microdevices, 2011, 13, 983–993 CrossRef CAS PubMed.
- K. T. Nguyen and J. L. West, Biomaterials, 2002, 23, 4307–4314 CrossRef CAS.
- J. L. Ifkovits and J. A. Burdick, Tissue Eng., 2007, 13, 2369–2385 CrossRef CAS PubMed.
- B. Baroli, J. Pharm. Sci., 2007, 96, 2197–2223 CrossRef CAS PubMed.
- G. A. Hudalla, T. S. Eng and W. L. Murphy, Biomacromolecules, 2008, 9, 842–849 CrossRef CAS PubMed.
- A. I. Van Den Bulcke, B. Bogdanov, N. De Rooze, E. H. Schacht, M. Cornelissen and H. Berghmans, Biomacromolecules, 2000, 1, 31–38 CrossRef CAS.
- J.-W. Choi, R. Wicker, S.-H. Lee, K.-H. Choi, C.-S. Ha and I. Chung, J. Mater. Process. Technol., 2009, 209, 5494–5503 CrossRef CAS PubMed.
- A. D. Lantada, P. L. Morgado and J. Stampfl, in Handbook on Advanced Design and Manufacturing Technologies for Biomedical Devices, Springer, US, 2013, pp. 181–205 Search PubMed.
- T. J. Horn and O. L. A. Harrysson, Sci. Prog., 2012, 95, 255–282 CrossRef CAS.
- S. D. Gittard and R. J. Narayan, Expert Rev. Med. Devices, 2010, 7, 343–356 CrossRef PubMed.
- A. Waldbaur, H. Rapp, K. Lange and B. E. Rapp, Anal. Methods, 2011, 3, 2681–2716 RSC.
- H. Lin, D. Zhang, P. G. Alexander, G. Yang, J. Tan, A. W. Cheng and R. S. Tuan, Biomaterials, 2013, 34, 331–339 CrossRef CAS PubMed.
- R. Liska, M. Schuster, R. Inführ, C. Turecek, C. Fritscher, B. Seidl, V. Schmidt, L. Kuna, A. Haase, F. Varga, H. Lichtenegger and J. Stampfl, J. Coat. Technol. Res., 2007, 4, 505–510 CrossRef CAS.
- L. H. Han, G. Mapili, S. Chen and K. Roy, J. Manuf. Sci. E–T. ASME, 2008, 130, 021005 CrossRef.
- P. Soman, J. A. Kelber, J. W. Lee, T. N. Wright, K. S. Vecchio, R. L. Klemke and S. Chen, Biomaterials, 2012, 33, 7064–7070 CrossRef CAS PubMed.
- P. Soman, B. T. D. Tobe, J. W. Lee, A. Winquist, I. Singec, K. S. Vecchio, E. Y. Snyder and S. Chen, Biomed. Microdevices, 2012, 14, 829–838 CrossRef CAS PubMed.
- P. Soman, D. Y. Fozdar, J. W. Lee, A. Phadke, S. Varghese and S. Chen, Soft Matter, 2012, 8, 4946–4951 RSC.
- P. Soman, J. W. Lee, A. Phadke, S. Varghese and S. Chen, Acta Biomater., 2012, 8, 2587–2594 CrossRef CAS PubMed.
- A. P. Zhang, X. Qu, P. Soman, K. C. Hribar, J. W. Lee, S. Chen and S. He, Adv. Mater., 2012, 24, 4266–4270 CrossRef CAS PubMed.
- D. Y. Fozdar, P. Soman, J. W. Lee, L. H. Han and S. Chen, Adv. Funct. Mater., 2011, 21, 2712–2720 CrossRef CAS PubMed.
- D. E. Discher, P. Janmey and Y.-L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- R. H. Baughman, S. Stafstram, C. Cui and S. O. Dantas, Science, 1998, 279, 1522–1524 CrossRef CAS.
- R. Lakes, Science, 1987, 235, 1038–1040 CAS.
- A. Khademhosseini and R. Langer, Biomaterials, 2007, 28, 5087–5092 CrossRef CAS PubMed.
- D. J. Odde and M. J. Renn, Biotechnol. Bioeng., 2000, 67, 312–318 CrossRef CAS.
- L. Koch, M. Gruene, C. Unger and B. Chichkov, Curr. Pharm. Biotechnol., 2013, 14, 91–97 CAS.
- R. S. Nathan, T. C. David, H. Yong, R. Nurazhani Abdul, X. Yubing and B. C. Douglas, Biofabrication, 2010, 2, 032001 CrossRef PubMed.
- P. K. Wu, B. R. Ringeisen, J. Callahan, M. Brooks, D. M. Bubb, H. D. Wu, A. Piqué, B. Spargo, R. A. McGill and D. B. Chrisey, Thin Solid Films, 2001, 398–399, 607–614 CrossRef CAS.
- A. Doraiswamy, R. J. Narayan, M. L. Harris, S. B. Qadri, R. Modi and D. B. Chrisey, J. Biomed. Mater. Res., Part A, 2007, 80, 635–643 CrossRef CAS PubMed.
- M. O. Christina, W. Xingjia, J. A. Juanita and R. R. Bradley, Biomed. Mater., 2008, 3, 034101 CrossRef PubMed.
- M. Gruene, M. Pflaum, C. Hess, S. Diamantouros, S. Schlie, A. Deiwick, L. Koch, M. Wilhelmi, S. Jockenhoevel, A. Haverich and B. Chichkov, Tissue Eng., Part C, 2011, 17, 973–982 CrossRef PubMed.
- L. Koch, A. Deiwick, S. Schlie, S. Michael, M. Gruene, V. Coger, D. Zychlinski, A. Schambach, K. Reimers, P. M. Vogt and B. Chichkov, Biotechnol. Bioeng., 2012, 109, 1855–1863 CrossRef CAS PubMed.
- R. K. Pirlo, P. Wu, J. Liu and B. Ringeisen, Biotechnol. Bioeng., 2012, 109, 262–273 CrossRef CAS PubMed.
- V. Chan, P. Zorlutuna, J. H. Jeong, H. Kong and R. Bashir, Lab Chip, 2010, 10, 2062–2070 RSC.
- F. P. W. Melchels, J. Feijen and D. W. Grijpma, Biomaterials, 2010, 31, 6121–6130 CrossRef CAS PubMed.
- D. W. Hutmacher, M. Sittinger and M. V. Risbud, Trends Biotechnol., 2004, 22, 354–362 CrossRef CAS PubMed.
- A. Ovsianikov, M. Gruene, M. Pflaum, L. Koch, F. Maiorana, M. Wilhelmi, A. Haverich and B. Chichkov, Biofabrication, 2010, 2, 014104 CrossRef CAS PubMed.
- G. Mapili, Y. Lu, S. C. Chen and K. Roy, J. Biomed. Mater. Res., Part B, 2005, 75, 414–424 CrossRef PubMed.
- M. Gebinoga, J. Katzmann, U. Fernekorn, J. Hampl, F. Weise, M. Klett, A. Läffert, T. A. Klar and A. Schober, Eng. Life Sci., 2013, 13, 368–375 CrossRef CAS.
- R. Landers, A. Pfister, U. Hubner, H. John, R. Schmelzeisen and R. Mulhaupt, J. Mater. Sci., 2002, 37, 3107–3116 CrossRef CAS.
- A. Ovsianikov, A. Deiwick, S. Van Vlierberghe, P. Dubruel, L. Moller, G. Dräger and B. Chichkov, Biomacromolecules, 2011, 12, 851–858 CrossRef CAS PubMed.
- E. T. Ritschdorff and J. B. Shear, Anal. Chem., 2010, 82, 8733–8737 CrossRef CAS PubMed.
- W. Zhang and S. C. Chen, MRS Bull., 2011, 1028–1033 CrossRef CAS.
- S. H. Lee, J. J. Moon and J. L. West, Biomaterials, 2008, 29, 2962–2968 CrossRef CAS PubMed.
- W. Zhang, P. Soman, K. Meggs, X. Qu and S. Chen, Adv. Funct. Mater., 2013, 23, 3226–3232 CrossRef CAS PubMed.
- S. Wu, J. Serbin and M. Gu, J. Photochem. Photobiol., A, 2006, 181, 1–11 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |



