Resveratrol and its metabolites inhibit pro-inflammatory effects of lipopolysaccharides in U-937 macrophages in plasma-representative concentrations
Jessica
Walker
*,
Katharina
Schueller
,
Lisa-Marie
Schaefer
,
Marc
Pignitter
,
Laura
Esefelder
and
Veronika
Somoza
University of Vienna, Department of Nutritional and Physiological Chemistry, Althanstrasse 14, A-1090 Vienna, Austria. E-mail: Jessica.Walker@univie.ac.at; Fax: +43-1-4277-9706; Tel: +43-1-4277-70611
First published on 13th November 2013
Abstract
Resveratrol has been shown to exploit various biological activities, including an anti-inflammatory activity. However, resveratrol is metabolized by phase II enzymes post-absorption to predominantly form glucuronides and sulfates. To investigate the anti-inflammatory effects of resveratrol and its dominating sulfated and glucuronated metabolites formed in vivo, U-937 macrophages were chosen as an immune-competent model system, known to release cytokines upon lipopolysaccharide stimulation. U-937 cells were stimulated with lipopolysaccharides from Escherichia coli (E. coli-LPS) to evoke an inflammatory reaction, and pre- or co-incubated with 1 or 10 μM of resveratrol (RES), resveratrol-3-sulfate (R3S), resveratrol-disulfates (RDS), resveratrol-3-glucuronide or resveratrol-4′-glucuronide. Time dependent gene expression of IL-6, IL-1α/β and IL-1R by qPCR was studied at 1 h, 3 h, 6 h, 9 h, and 24 h of incubation, and the release of IL-6 and TNF-α, after 6 h was analysed by means of non-magnetic or magnetic bead analysis. As a result, 10 μM resveratrol completely inhibited the E. coli-LPS-induced release of IL-6, while resveratrol-3-sulfate and resveratrol-disulfates decreased it by respective 84.2 ± 29.4% and 52.3 ± 39.5%. Whereas TNF-α release was reduced by 48.1 ± 15.4%, 33.0 ± 10.0% and 46.7 ± 8.7% by RES, R3S and RDS, respectively. These results show that not only resveratrol but also resveratrol-3-sulfate and resveratrol-disulfates exhibit an anti-inflammatory potential by counteracting an inflammatory challenge in U-937 macrophages at plasma representative concentrations.
Introduction
Resveratrol, a stilbenoid, is commonly found in peanuts, grapes and red wine.1,2 It has been identified to possess antioxidant,3 chemopreventive4,5 and anti-inflammatory activities.6 However, upon digestion and absorption, resveratrol is metabolized by phase II enzymes, resulting mainly in the formation of sulfated and glucuronated metabolites.7–10 In human intervention studies,8,10,11 the following metabolites have been identified in the human plasma and urine: resveratrol-3-sulfate (Res-3-sulf, R3S), resveratrol-3,5-disulfate, resveratrol-3,4′-disulfate (Res-disulfates, RDS), resveratrol-4′-sulfate, resveratrol-3-glucuronide (Res-3-gluc, R3G) and resveratrol-4′-glucuronide (Res-4′-gluc, R4′G). After intake of a bolus dose of 85.5 mg per 70 kg body weight of piceid, a resveratrol glucoside, the in 3-position monosulfated metabolite and the disulfated metabolites were the predominant metabolites detected after 6 h in the plasma of the volunteers, reaching a maximum concentration of 1 μM.8 In addition, after consumption of a high bolus of 5 g resveratrol, resveratrol-3-sulfate was detected as the most abundant metabolite with a concentration of up to 14 μM in the plasma. In the same study, resveratrol sulfates and glucuronides were analyzed in the plasma with half-lives between 3.2 and 11.5 h, and 2.9 to 10.6 h, respectively.11 Moreover, an uptake of repeated doses of up to 5 g resveratrol per day for 21 days resulted in a maximal plasma concentration of about 13 μM resveratrol-3-sulfate.12 Incubation times and the concentrations used in in vitro studies focusing on the biological effects of these metabolites should, therefore, not exceed 14 μM.The effects of resveratrol on the release of cytokines after stimulation with lipopolysaccharides (LPS) have been studied in various cell models. In a study using Caco-2 cells, a human colon cancer cell-line, it has been shown that resveratrol, up to a concentration of 50 μM, inhibits the secretion of the pro-inflammatory mediators prostaglandin E2 and cyclooxygenase-2, when treated with the compound for 1 h prior to a 48 h challenge with LPS. Furthermore, resveratrol has been shown to elicit an anti-inflammatory activity in macrophages by reducing the LPS-stimulated release of interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α), gene expression of IL-1β in concentrations between 1 and 10 μM after 6 h of incubation.13
Gram-negative bacteria, like Escherichia coli (E. coli), express the endotoxins LPS on their surface, which evoke an inflammatory reaction of the host when offered to the immune system.14,15 Rittig and co-workers16 showed by measurement of different cytokines that the immune response to LPS varies depending on the LPS phenotype. A treatment of human peripheral blood mononuclear cells with media from platelets which were stimulated with LPS from E. coli led to a release of the interleukins-6 (IL-6) and -8 (IL-8), as well as the matrix metalloproteases-2 (MMP-2) and -9 (MMP-9), with a different efficacy than a treatment of the platelets with LPS from Salmonella minnesota.17 However, E. coli are ubiquitous bacteria that can cause food poisoning as well as sepsis. Therefore, LPS from E. coli should be used analyzing a pro-inflammatory response in blood, monocytes or macrophages. Taken into account that different types of LPS may affect the release of cytokines from one cell model to another with different efficacies, prior to our investigation of the influence of resveratrol on the immune response, the optimal marker for the respective test system, considering the type of LPS and the cell-line used, needed to be identified. Of particular interest for the identification of immune-modulatory effects are immune-competent cells, like monocytes or macrophages. For in vitro studies, U-937 macrophages are widely studied since these cells grow as monocytes in suspension and have been demonstrated to release IL-6 after stimulation with IL-1β,18 and MMP-2 and MMP-9 after stimulation with E. coli-LPS.19 The U-937 monocytes can be differentiated with phorbol-12-myristate-13-acetate (PMA) into U-937 macrophages that may serve as a model system to identify an anti-inflammatory activity of natural compounds by means of a reduction of TNF-α, IL-6 and IL-1β gene expression.20 In our study, we first aimed to determine the optimal cell culture conditions to study the anti-inflammatory potential of a target compound after incubation of U-937 macrophages with LPS from E. coli. Afterwards, we chose E. coli-LPS-stimulated U-937 macrophages as an in vitro system to elucidate the anti-inflammatory effects of plasma representative concentrations of resveratrol sulfates and glucuronides in comparison to their parent compound, since data on the anti-inflammatory potential of the resveratrol plasma metabolites are limited to the use of concentrations ≥30 μM and incubation times of up to 48 h.21,22
Materials and methods
Materials
All chemicals were purchased from Sigma-Aldrich (Vienna, Austria), unless indicated otherwise. Lipopolysaccharide from E. coli (055:B5) was reconstituted to a concentration of 10 mg mL−1 and aliquots were stored at −20 °C. The primers designed (primer3) were verified with primer BLAST (NCBI) and synthesized by VBC-Biotech (Vienna, Austria) (Table 1).| Primer | Direction | Sequence | T m [°C] |
|---|---|---|---|
| PPIA | Sense | 5′-CCA CCA GAT CAT TCC TTC TGT AGC-3′ | 55.8 |
| Anti-sense | 5′-CTG CAA TCC AGC TAG GCA TGG-3′ | 57.1 | |
| IL-6 | Sense | 5′-AAA CAA CCT GAA CCT TCC AAA GA -3′ | 57.1 |
| Anti-sense | 5′-GCA AGT CTC CTC ATT GAA TCC A -3′ | 58.4 | |
| IL-6R | Sense | 5′-CCA CCC CCA TGC AGG CAC TTA-3′ | 61.4 |
| Anti-sense | 5′-CCA GGC TCC CTC CAG CAA CCA-3′ | 63.0 | |
| IL-1α | Sense | 5′-CCA GTG CTG CTG AAG GAG ATG-3′ | 61.9 |
| Anti-sense | 5′-AGT GCC GTG AGT TTC CCA GAA-3′ | 58.8 | |
| IL-1β | Sense | 5′-GGA GAA CAC CAC TTG TTG CAC CAT-3′ | 60.2 |
| Anti-sense | 5′-ACA GAG CCT CGC CTT TGC CG-3′ | 58.1 | |
| IL-1R | Sense | 5′-CCC TCC TGA GAA GCT GGA CCC C-3′ | 61.9 |
| Anti-sense | 5′-CGT TCC TTG CAT TCA GCC TCC A-3′ | 62.0 | |
| TNF-α | Sense | 5′-CCC ATG TTG TAG CAA ACC CTC AA-3′ | 68.7 |
| Anti-sense | 5′-TAT CTC TCA GCT CCA CGC CAT T-3′ | 67.0 |
Trans-resveratrol metabolites were synthesized as described in previous publications of our group.8,9,23 The structure and purity of the synthesized compounds are shown in Fig. 1. Resveratrol and the resveratrol metabolites were dissolved in 100% DMSO to a concentration of 10 mM. These stock solutions were prepared freshly prior to the experiments and kept in the dark during handling procedures.
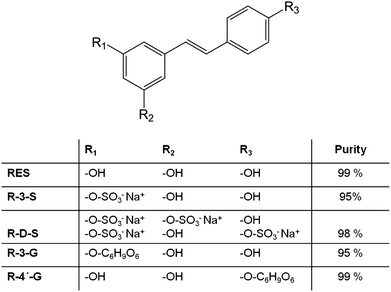 | ||
| Fig. 1 Structure of t-resveratrol (RES), t-resveratrol-3-sulfate (R-3-S), t-resveratrol-disulfate (R-D-S), t-resveratrol-3-glucuronide (R-3-G) and t-resveratrol-4′-glucuronide (R-4′-G) and purity9 of synthesized compounds. | ||
U-937 cell culture and differentiation procedure
The U-937 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, U.S.A.), cultured in Roswell Park Memorial Institute 1640 (RPMI) medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Vienna, Austria), 100 U ml−1 penicillin and 0.1 mg ml−1 streptomycin (Invitrogen) in a humidified incubator with 5% CO2 at 37 °C, and split every 2–3 days. To differentiate the U-937 monocytes into macrophages, the cells were seeded in the respective volume and plate or dish in RPMI media containing 10% FBS, 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin, 2 mM glutamine and 10 or 50 ng ml−1 phorbol-12-myristate-13-acetate (PMA). After 48 h, the differentiated cells were used for the experiments. The differentiation was verified by detection of CD11b using flow cytometry.24 The cells were incubated with anti-CD11b antibody (sc-1186, Integrin αM, Santa Cruz Biotechnologies, Heidelberg, Germany) for 1 h and a fluorescein isothiocyanate (FITC)-coupled secondary antibody (sc-2078, goat anti-mouse, IgG1-FITC, Santa Cruz Biotechnologies) for 30 min or a control antibody (sc-2855, normal mouse IgG1-FITC, Santa Cruz Biotechnologies) for 1 h. The FITC signal was measured using flow cytometry (guava, Merck-Millipore, Vienna, Austria) and the data was analyzed with the InCyte software (v 2.2.2, Merck-Millipore).U-937 macrophage cell viability and incubation media sampling
The viability of the U-937 macrophages was tested by trypan blue staining. The cells were seeded in 24-well plates with 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and differentiated as described above. The differentiation media was exchanged with media containing 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin, 2 mM glutamine, 10% FBS, and the test compounds and incubated for 3, 6, 9 or 24 h, respectively. To test a preventive effect of the compounds the cells were incubated for 1 h with the respective compound, followed by 6 h stimulation with E. coli-LPS in absence of the compound. For resveratrol experiments, 1% DMSO was used as solvent control and 100 nM dexamethasone was used as an anti-inflammatory active positive control compound.25,26 After incubation, the incubation media was collected for cytokine analyses and stored at −80 °C. The cells were detached with 0.2% EDTA in PBS. The cell suspension was diluted 1
000 cells per well and differentiated as described above. The differentiation media was exchanged with media containing 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin, 2 mM glutamine, 10% FBS, and the test compounds and incubated for 3, 6, 9 or 24 h, respectively. To test a preventive effect of the compounds the cells were incubated for 1 h with the respective compound, followed by 6 h stimulation with E. coli-LPS in absence of the compound. For resveratrol experiments, 1% DMSO was used as solvent control and 100 nM dexamethasone was used as an anti-inflammatory active positive control compound.25,26 After incubation, the incubation media was collected for cytokine analyses and stored at −80 °C. The cells were detached with 0.2% EDTA in PBS. The cell suspension was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with trypan blue solution (0.4%) and the cell viability was determined using a hemocytometer with n = 3 and four technical replicates.
10 with trypan blue solution (0.4%) and the cell viability was determined using a hemocytometer with n = 3 and four technical replicates.
MMP-2 ELISA
The cell culture media of the 24 h incubation were thawed and used undiluted in the MMP-2 ELISA (Calbiochem/Millipore, Vienna, Austria), following the manufacture's protocol. The absorbance was measured at 450 nm against a reference of 595 nm using a multimode plate reader (Tecan infinite M2000, Tecan, Groeding, Austria). Three independent experiments were carried out and duplicates of each sample were measured with the ELISA kit. The concentration of MMP-2 in the cell culture media was determined using a standard curve. All samples were compared to the E. coli-LPS treated positive control (100%) and are presented as T/LPS (%).Cytokine measurement via bead-kit
The cytokines IL-6, IL-8 and MMP-9 were determined in the thawed incubation media using non-magnetic or magnetic bead kits (both ebioscience, Vienna, Austria).27 For the measurement of MMP-9 the media had to be diluted to stay within the standard range depending on the incubation time since concentrations of MMP-9 increased over time, for 3 h 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, for 6 h 1
3, for 6 h 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, for 9 h 1
5, for 9 h 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and for 24 h 1
10 and for 24 h 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. Three to four independent experiments were carried out and duplicates of each sample were analyzed within the bead kit measurement following the manufacture's protocols. The detection of the non-magnetic beads was carried out on a flow cytometer (Guava, Merck-Millipore), with a bead emission at 700 nm and the phycoerythrin emission at 575 nm. Data were analyzed using the FlowCytoMix Pro software (ebioscience). For the detection and analysis using magnetic beads the MagPix system and Milliplex analyst software (Merck-Millipore) were used, respectively. The bead regions were gated and based on standard curves the results were calculated as pg ml−1. The results were normalized to the stimulated control, 1 μg ml−1E. coli-LPS (100%), as T/LPS in %.
50. Three to four independent experiments were carried out and duplicates of each sample were analyzed within the bead kit measurement following the manufacture's protocols. The detection of the non-magnetic beads was carried out on a flow cytometer (Guava, Merck-Millipore), with a bead emission at 700 nm and the phycoerythrin emission at 575 nm. Data were analyzed using the FlowCytoMix Pro software (ebioscience). For the detection and analysis using magnetic beads the MagPix system and Milliplex analyst software (Merck-Millipore) were used, respectively. The bead regions were gated and based on standard curves the results were calculated as pg ml−1. The results were normalized to the stimulated control, 1 μg ml−1E. coli-LPS (100%), as T/LPS in %.
Gene expression analysis of cytokines and cytokine receptors
For gene expression analysis of IL-1α, IL-1β, IL-1R, IL-6, IL-6R and TNF-α, PPIA (Table 1) was used as a reference gene.28,29 Five million U-937 cells were seeded in cell culture dishes (10 cm diameter) and differentiated for 48 h. Thereafter, the cells were incubated for 3, 6, 9 or 24 h with the respective compounds in combination with 1 μg ml−1E. coli-LPS. After washing the cells twice with ice-cold PBS, the RNA was isolated using the SV RNA total isolation kit (Promega) and RNA concentration and purity were analyzed spectrophotometrically using a NanoQuant plate® (Tecan). The purities of all samples were measured with ratios at 260/280 nm between 1.8 and 2.3. Of each sample, 2 μg of RNA were transcribed into cDNA using the high capacity reverse transcription kit with RNase inhibitor (Life Technologies). The cDNA concentration and purity was determined with a NanoQuant plate® (Tecan). The qPCR was carried out on a StepOnePlus instrument (ABI) using 50 ng of cDNA and 200 nM of the specific primer pairs (Table 1). For data analysis, the PCR efficiencies and corrected N0-values were determined using LinRegPCR.28 The N0-values, the starting concentrations, of each sample were normalized to the reference gene, PPIA. The fold changes of the N0-values from E. coli-LPS-treated versus the untreated control cells were calculated and displayed for the analysis of time-dependent effects of E. coli-LPS stimulation on gene expression. The influence of resveratrol and resveratrol metabolites were compared to the E. coli-LPS treatment (100%) and presented as T/LPS control (%).Statistical analysis
Three to four independent experiments were carried out for all analyses. In all experiments, outliers were excluded using a Nalimov outlier test with p < 0.05. The effects of PMA differentiation, E. coli-LPS stimulation and resveratrol and its metabolites on the LPS stimulation were tested using a two sided Student's t-test. Significant differences are displayed with *p < 0.05, **p < 0.01 and ***p < 0.001. For the determination of the optimal incubation time and the effects of the different substances, a one-way ANOVA with a Holm–Sidak post-hoc test was performed between the different incubation times for each gene and each concentration. Significant differences (p < 0.05) are indicated with letters.Results
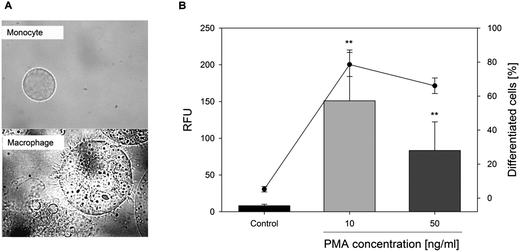
Differentiation of U-937 monocytes into macrophages
The differentiation of U-937 monocytes into macrophages was initiated with phorbol-12-myristate-13-acetate (PMA).30 The optimal PMA concentration was determined using integrin alpha M (cluster of differentiation 11b, CD11b) presentation capacity on the cell surface by flow cytometry and, additionally, the cell morphology was checked by microscopy (Fig. 2A). CD11b, one subunit of the heterodimeric integrin alpha M beta 2 receptor, has been established to determine the differentiation of macrophages.31 Treatment of U-937 monocytes with 10 ng ml−1 PMA for 48 h led to differentiation of 78.6 ± 7.1%, whereas a treatment with 50 ng ml−1 PMA for the same incubation time did not result in a higher proportion of differentiated cells (Fig. 2B). Thus, for all further experiments, the monocytes were differentiated into macrophages by treatment with 10 ng ml−1 PMA for 48 h. For the experiments, the remaining monocytes in suspension were removed by washing with PBS and changing of media to incubation media, thus, only macrophages were used in the experimental setting.Determination of the optimal incubation time for cytokine gene expression and cytokine release after stimulation of the U-937 macrophages with E. coli-LPS
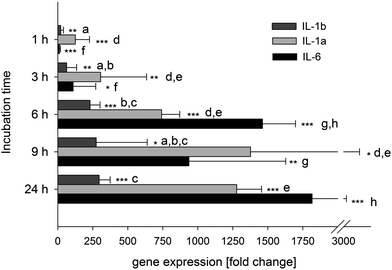
The gene expression of IL-6 as well as IL-1α, IL-1β, IL-1R and IL-6R was measured after incubation with and without 1 μg ml−1E. coli-LPS for 1, 3, 6, 9 and 24 h. At all times, the incubation of the differentiated U-937 cells with 1 μg ml−1E. coli-LPS had no cytotoxic effect compared to the untreated control (data not shown). A strong increase in the expression of IL-1α, IL-1β and IL-6 to a fold change of 742 ± 130, 230 ± 72.5 and 1462 ± 235, respectively, was determined after 6 h of stimulation (Fig. 3). At the later time points, the increase was not as pronounced. The IL-1 receptor gene was up-regulated by E. coli-LPS stimulation for 6 h to a fold change of 4.92 ± 1.27, while IL-6R was down-regulated to 0.86 ± 0.08 (data not shown). Taken together, we chose the 6 h incubation time to measure the influence of resveratrol and its metabolites on the gene expression in further experiments.To determine the optimal incubation time for the cytokine and matrix metalloprotease (MMP) release by stimulation of U-937 macrophages with 1 μg ml−1E. coli-LPS,32 the time dependent release of IL-6, IL-8, and MMP-9 was measured at 3, 6, 9 and 24 h. IL-6, IL-8 and MMP-9 secretion after 3, 6, 9 and 24 h incubation were measured in the cell culture media and the data is represented in Table 2 as fold change of the E. coli-LPS treatment vs. untreated control cells. The higher this value the more effective was the E. coli-LPS stimulation. After 3 h, only IL-8 was stimulated to a fold change of 1.74 ± 0.43. However, this stimulation was not detectable after 6, 9 or 24 h. IL-6 release increased over time and was stimulated after exposure times of 6 h and longer. In contrast to the MMP-2 release, the MMP-9 release was stimulated after 24 h. After 6 h, IL-6 gene expression was stimulated to a maximum and also IL-6 release was significantly increased compared to the control. Finally, IL-6 was chosen as a biomarker in the E. coli-LPS-stimulated U-937 macrophages to determine the influence of resveratrol and its metabolites in the further experiments. In addition, the release of TNF-α, known to be up-regulated on a transcriptional level33 and released from U-937 cells after LPS stimulation,34,35 was studied. An increased mRNA expression and release of this marker after 6 h of stimulation with 1 μg ml−1E. coli-LPS of 14.3 ± 9.0 fold and 314.9 ± 54.1 fold, respectively, compared to the non-stimulated control was determined (data not shown).
| IL-6 | IL-8 | MMP-9 | MMP-2 | |||||
|---|---|---|---|---|---|---|---|---|
| 3 h | 1.80 ± 2.07 | 1.74 ± 0.43 | ** | 0.90 ± 0.54 | n.d. | |||
| 6 h | 13.0 ± 1.87 | *** | 0.85 ± 0.13 | 0.94 ± 0.34 | n.d. | |||
| 9 h | 58.6 ± 22.0 | *** | 0.49 ± 0.02 | *** | 1.63 ± 0.32 | n.d. | ||
| 24 h | 207 ± 45.0 | *** | 0.32 ± 0.03 | ** | 3.17 ± 0.28 | *** | 0.55 ± 0.07 | *** |
Inhibition of the expression of pro-inflammatory genes by resveratrol and its metabolites
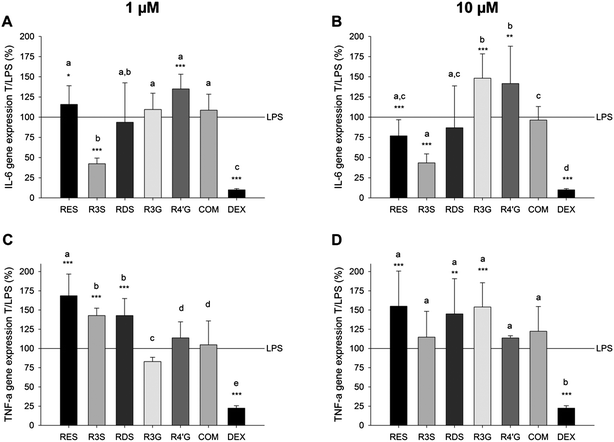
The impact of resveratrol, resveratrol sulfates and glucuronides on the expression of IL-6, TNF-α, IL-1α, IL-1β and the IL-1R in U-937 macrophages was determined after a 6 h co-incubation with E. coli-LPS (Table 3 and Fig. 4). Dexamethasone in a concentration of 100 nM was used as an anti-inflammatory control, up-regulating IL-1R mRNA expression to 344 ± 16% and down-regulating the gene expression of IL-1α, IL-1β (data not shown), IL-6 and TNF-α (Figure 4) to 5.6 ± 0.7%, 27.9 ± 2.7%, 9.9 ± 1.6% and 22.4 ± 3.1%, respectively, compared to the E. coli-LPS treatment solely (100%).| Conc. | IL-1α | IL-1β | ||
|---|---|---|---|---|
| 1 μM | 10 μM | 1 μM | 10 μM | |
| RES | 49.7 ± 10.0*a | 48.0 ± 4.9*a | 77.0 ± 40.0a,b | 79.1 ± 15.1*a,c |
| R3S | 61.2 ± 17.0*a | 45.5 ± 15.5*a | 76.6 ± 29.5a | 58.4 ± 16.7*a |
| RDS | 55.3 ± 25.2*a | 79.9 ± 41.2*a,b | 59.8 ± 33.6*a | 76.0 ± 34.2*a,c |
| R3G | 86.5 ± 10.6*b | 111 ± 31.2*b | 102 ± 14.1b,c | 127 ± 12.0*b |
| R4′G | 111 ± 14.1c | 119 ± 52.3*b | 128 ± 32.6*c | 127 ± 20.4*b |
| COM | 89.4 ± 3.95*b | 117 ± 13.0*b | 90.2 ± 6.14*b | 94.8 ± 7.60*c |
| DEX 100 nM | 5.56 ± 0.74* | 27.9 ± 2.67* | ||
Next, the impact of resveratrol and resveratrol metabolites on these target genes was investigated. Prior to the gene expression experiments using resveratrol or resveratrol metabolites in combination with LPS, false positive results due to cytotoxic effects had to be excluded. Co-incubation of 1 μg ml−1E. coli-LPS and 10 μM resveratrol, resveratrol-3-sulfate, resveratrol disulfates, resveratrol-3-glucuronide or resveratrol-4′-glucuronide did not show cytotoxic effects compared to the solvent control (data not shown).
Resveratrol, at a concentration of 10 μM, suppressed the LPS-stimulated expression of IL-1α and IL-1β to 48.0 ± 4.9% and 79.1 ± 15.0%, respectively (Table 3). The effects of the tested resveratrol-3-sulfate and resveratrol disulfates concentrations on the gene expression of IL-1α and IL-1β did not differ from the inhibitory effect of resveratrol, and reduced the expression of these genes to a maximum of 45.5 ± 15.5% and 58.4 ± 16.7%, respectively. In contrast to the sulfated metabolites, the tested resveratrol glucuronides mostly stimulated the gene expression of the tested genes by additional 8.6 ± 15.6% to 48.2 ± 30.1% compared to the LPS treatment solely. Only 1 μM resveratrol-3-glucuronide slightly inhibited the expression of IL-1α and IL-1R to 86.5 ± 10.6% and 80.0 ± 9.7%, respectively.
The gene expression of IL-6 was most effectively reduced to 42.2 ± 11.2% in the presence of 1 μM resveratrol-3-sulfate compared to resveratrol and the other metabolites (Fig. 4). However, at a concentration of 10 μM, resveratrol and the sulfated metabolites exploited similar anti-inflammatory activity by reducing the E. coli-LPS-stimulated IL-6 expression to a maximum of 43.4 ± 11.1% (R3S). In contrast, the TNF-α mRNA expression was up-regulated after co-incubation with 1 or 10 μM resveratrol and 1 μg ml−1E. coli-LPS for 6 h. Similar effects were found for the sulfated metabolites, while the glucuronated metabolites did not influence the gene expression of TNF-α in comparison to a treatment with LPS solely.
Taken together, with regard to the expression of the majority of tested pro-inflammatory genes, the resveratrol sulfates exhibited a strong anti-inflammatory potential, while resveratrol monoglucuronides did not. Thus, mixtures of either 1 or 10 μM of each of the metabolites were tested as well to prove the combined effect of resveratrol metabolites. This mixture (COM) did not impact the IL-6 or TNF-α mRNA expression (Fig. 4), but reduced the pro-inflammatory effect of E. coli-LPS by means of IL-1α and IL-1β mRNA expression levels (Table 3).
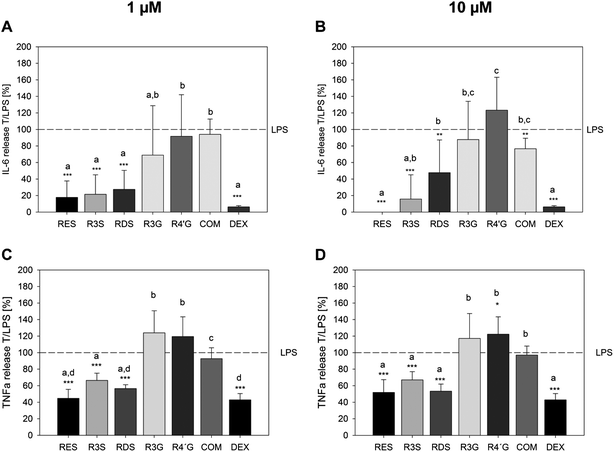
Resveratrol and resveratrol metabolites inhibit the IL-6 and TNF-α release of from U-937 macrophages after 6 h of incubation
On a functional level, it was analyzed whether resveratrol and its metabolites could inhibit the E. coli-LPS stimulated IL-6 and TNF-α release from U-937 macrophages. The anti-inflammatory control compound dexamethasone decreased the release of IL-6 and TNF-α, when co-incubated at a concentration of 100 nM together with 1 μg ml−1E. coli-LPS for 6 h, by 93.7 ± 1.52% and 57.2 ± 7.69%, respectively, compared to the treatment with LPS solely (Fig. 5). Resveratrol strongly inhibited the pro-inflammatory response of E. coli-LPS by means of IL-6 release already at a concentration of 1 μM (17.8 ± 19.8%, p < 0.001, Fig. 5A) and completely at a concentration of 10 μM (Fig. 5B). This anti-inflammatory activity of resveratrol was also shown with regards to the LPS-stimulated TNF-α release. Here, 1 μM resveratrol inhibited the endotoxin-mediated cytokine release to 44.7 ± 11.0% (Fig. 5C). In addition, the sulfated metabolites showed an inhibition of the LPS-stimulated IL-6 and TNF-α release after 6 h of incubation. Here, the monosulfated metabolite tested suppressed the IL-6 release to a minimum of 15.8 ± 29.4% at 10 μM and the TNF-α release to a minimum of 66.4 ± 8.7% at 1 μM, while the disulfated metabolites reduced the IL-6 release to 27.5 ± 23.0% at 1 μM and the TNF-α release to 53.3 ± 8.7% at 10 μM. In contrast, the glucuronated metabolites did not inhibit the E. coli-LPS induced IL-6 or TNF-α release from U-937 macrophages. A concomitant incubation of 1 μg ml−1E. coli-LPS and a combination 10 μM of each of the metabolites resulted in a reduction of the LPS-stimulated IL-6 release (Fig. 5B), but did not influence the TNF-α release (Fig. 5D).Resveratrol metabolites exhibit a preventive effect against a challenge of U-937 macrophages stimulated with E. coli-LPS
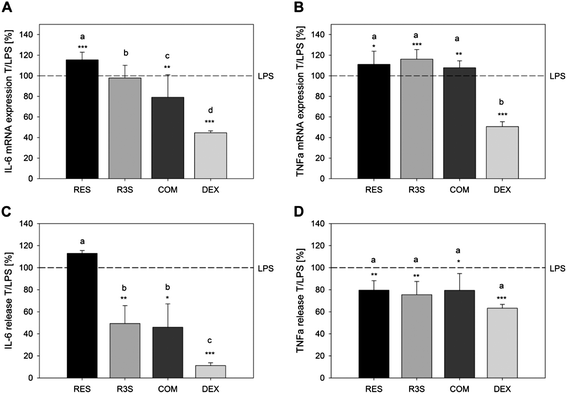
In addition to the effect of resveratrol and its metabolites concomitantly incubated with LPS, a preventive effect of the most active substances and the metabolite mixture against the pro-inflammatory activity of E. coli-LPS was investigated by incubation of U-937 cells with 10 μM resveratrol, resveratrol-3-sulfate or the combination of 10 μM of the resveratrol metabolites for 1 h, followed by a six hour stimulation with 1 μg ml−1E. coli-LPS. Dexamethasone served as an anti-inflammatory control, up-regulating the LPS-stimulated IL-1R mRNA expression and down-regulating the gene expression of IL-1α, IL-1β, IL-6 and TNF-α (Fig. 6A and B), as well as reducing the LPS-mediated release (Fig. 6C and D) of the pro-inflammatory cytokines IL-6 by 88.8 ± 2.4% and TNF-α by 46.8 ± 3.4% compared to the control (1 h 1% DMSO + 6 h 1 μg ml−1E. coli-LPS). Resveratrol did not inhibit the up-regulation of pro-inflammatory cytokine mRNA expression by LPS. In contrast, a pre-incubation with a mixture of 10 μM of each of the metabolites resulted in a decreased IL-6 mRNA expression (79.1 ± 22.0, Fig. 6A) compared to the DMSO pre-treated control (100%). On a functional level, resveratrol also showed no inhibition of the E. coli-LPS-stimulated IL-6 release (Fig. 6C) but reduced the TNF-α release by 20.4 ± 8.65% (Fig. 6D). The tested sulfated metabolite and the metabolite mixture showed comparable effects on the LPS-mediated TNF-α release. However, a 1 h pre-treatment of the macrophages with resveratrol-3-sulfate or the metabolite mixture followed by a 6 h LPS-stimulation resulted in an inhibition of the IL-6 release to a minimum of 49.3 ± 16.2% and 46.0 ± 21.2%, respectively. Thus, resveratrol-3-sulfate and a mixture of tested resveratrol metabolites showed a greater preventive potential against the pro-inflammatory endotoxin E. coli-LPS than the parent compound.Discussion
Resveratrol is a known anti-inflammatory compound, but is metabolized upon absorption. We hypothesized that the resveratrol metabolites might also possess anti-inflammatory activities with regards to cytokine release in plasma representative concentrations. Since the release of cytokines depend on the type of LPS and cells used,16,17 prior to the analysis of an impact of resveratrol and its metabolites on the LPS-stimulated release of interleukins and matrix metalloproteases, the optimal marker needed to be identified. We determined IL-6 as well as TNF-α to be the ideal markers for our study by means of gene expression and interleukin secretion after 6 h of stimulation with 1 μg ml−1E. coli-LPS. In previous studies on U-937 monocytes, we measured the release of matrix metalloproteases (MMP-2 and MMP-9) after 24 h of incubation.19 In the here presented study, the release of MMP-9 could only be stimulated with 1 μg ml−1E. coli-LPS after 24 h. This result demonstrates that the release of matrix metalloproteases is a late event in the immune response, while the release of interleukins is an early one. The release of IL-6 was already significantly stimulated after 6 h. Furthermore, it is known that macrophages release TNF-α upon stimulation with LPS.34,35 Since resveratrol and its metabolites have been detected in the plasma with half lives of 2.9 to 11.5 h7,8 the treatment time with the compounds of interest should be kept within this time frame. With respect to the here presented study, the incubation time of six hours applied represent biologically relevant conditions and optimal conditions with regards to IL-6 and TNF-α release from U-937 macrophages.Resveratrol and resveratrol-3-sulfate have been shown to act as anti-inflammatory agents, affecting the expression of pro-inflammatory genes or inhibiting the release of pro-inflammatory cytokines. Furthermore, in U-937 macrophages, resveratrol was shown to down-regulate the gene expression of MMP-9.36 Our results on the inhibition of the E. coli-LPS induced IL-6 and TNF-α release and gene expression of U-937 macrophages are in accordance with a study from Qureshi et al.,37 showing that 0.1 to 10 μM resveratrol inhibit the LPS-stimulated release of TNF-α and gene expression of TNF-α, IL-1β, IL-6 and iNOS from RAW 264.7 macrophages.
Here, we could also show that not only resveratrol but also its sulfated metabolites exhibit anti-inflammatory activities by inhibiting the E. coli-LPS stimulated mRNA expression of pro-inflammatory cytokines and release of IL-6 and TNF-α by U-937 cells. In contrast to literature evidence for glucuronides of quercetin,38 the results of this study demonstrate that resveratrol glucuronides do not possess anti-inflammatory properties by an inhibition of IL-6 or TNF-α release from macrophages. Furthermore, resveratrol-3-sulfate inhibited the E. coli-LPS-stimulated gene expression of the pro-inflammatory genes IL-1α, IL-1β and IL-1R in a similar manner as resveratrol in plasma representative concentrations. Besides resveratrol, this metabolite was the most effective anti-inflammatory compound tested. These results are in accordance with the studies from Aires et al.,21 investigating the anti-proliferating activity of resveratrol metabolites, and Hoshino et al.22 who studied the anti-inflammatory activities of resveratrol sulfates in different test systems at higher concentrations. Both groups identified resveratrol-3-sulfate as the most active compound besides resveratrol. Aires et al.21 demonstrated that resveratrol-3-sulfate inhibits the proliferation of colon carcinoma cells (SW480, SW620) after 48 h in concentrations ≥10 μM. Also a concentration of 30 μM resveratrol-3-sulfate, the most active metabolite in our test system, was found to induce apoptosis in the same manner as resveratrol.21,39 Furthermore, Hoshino et al.22 studied the anti-inflammatory potential of sulfated resveratrol metabolites in RAW 264.7 macrophages with regards to their impact on the eicosanoid pathway and iNOS expression. Here, 34 μM resveratrol-3-sulfate were found to reduce the iNOS protein level, induced by 1 μg ml−1 LPS. In human intervention trials focusing on the metabolism of resveratrol, resveratrol sulfates and glucuronides have been identified in the plasma and urine of healthy volunteers, with resveratrol-3-sulfate being the predominant metabolite.8,10,11 However, in the previously mentioned studies by Aires et al.21 and Hoshino et al.22 in which the biological effects of resveratrol metabolites were investigated, the applied concentrations exceeded the plasma levels for resveratrol-3-sulfate by up to 2.4 times, based on an intake of a high bolus dose of resveratrol.11 Thus, for the first time, we demonstrated an anti-inflammatory activity for resveratrol metabolites in plasma representative concentrations.
In addition, we tested whether the plasma-representative concentrations of resveratrol, resveratrol-3-sulfate, the predominant metabolite, and a mixture of the metabolites exploit a preventive effect against a treatment with 1 μg ml−1E. coli-LPS. Here, we could demonstrate that resveratrol-3-sulfate and the mix of the tested metabolites attenuate the LPS-induced IL-6 and TNF-α release, after a 1 h treatment with the compound followed by 6 h LPS stimulation. Similar effects have been shown for lycopene40 using RAW264-7 macrophages and stimulation with LPS. In the same cell model, a 30 min treatment with resveratrol (≥25 μM) followed by an 8 h LPS stimulation resulted in a reduction of the IL-6 and TNF-α release compared to the LPS treatment solely.41 Our study shows, that the metabolites have a more pronounced effect under these conditions than the parent compound and exhibit their preventive potential against inflammation in plasma-representative concentrations.
Several studies focused on the identification of the mechanism of action. Resveratrol was found to block elements of the JAK/STAT-1 pathway in macrophages42 as well as the NF-κB signalling and the Toll-like receptor 4,41 resulting in a decreased mRNA level of cytokines and chemokines. For example, resveratrol inhibited the expression of the IL-6 gene.43 Additionally, an inhibition of the LPS-induced stimulation of the phosphatidylinositol 3-kinase pathway by resveratrol was recently shown, which led to a decreased expression of the genes encoding IL-1β, TNF-α, iNOS and cyclooxygenase-2.13 We were able to show that resveratrol-3-sulfate and the resveratrol disulfates decreased the LPS-induced gene expression of IL-1α, IL-1β and IL-6. Thus, the mechanism described, might apply to explain how resveratrol-3-sulfate and the resveratrol disulfates act as anti-inflammatory agents. However, the anti-inflammatory control, used in the present study, dexamethasone is known to inhibit the translocation of NF-κB to the nucleus and its binding to the DNA in macrophages leading to an attenuation of LPS-induced expression of pro-inflammatory cytokines.44 This result was clearly shown for all tested mRNA expression levels, here, while the IL-1R expression was up-regulated in the presence of LPS and dexamethasone. In contrast, resveratrol showed different effects on the IL-1R, IL-6 and TNF-α mRNA expression compared to dexamethasone. This could indicate a different mechanism of action by resveratrol. While dexamethasone influences the main inflammatory NF-κB pathway directly, resveratrol and resveratrol-3-sulfate act via different pathways (PI3K, JAK/STAT, NF-κB), with a lower impact on the resulting expression of pro-inflammatory genes. With special regards to the preventive effects, the mechanism of action of the metabolites should be investigated in future studies.
Conclusion
In conclusion, plasma representative concentrations of resveratrol sulfates possess anti-inflammatory activities, showing that resveratrol-3-sulfate, found in higher plasma concentrations than the parent compound, might contribute to its anti-inflammatory effect. However, only a small number of resveratrol metabolites were tested here. Future studies should elucidate the anti-inflammatory potential of other metabolites.Non-conflict of interest statement
The authors confirm that there is no conflict of interest with any financial organization regarding the results presented in this research manuscript.Abbreviations
| FITC | Fluorescein isothiocyanate |
| IL | Interleukin |
| iNOS | Inducible nitric oxide synthase |
| LPS | Lipopolysaccharides |
| MMP | Matrix metalloprotease |
| PMA | Phorbol-12-myristate-13-acetate |
| R3G | Resveratrol-3-glucuronide |
| R4′G | Resveratrol-4′-glucuronide |
| R3S | Resveratrol-3-sulfate |
| RDS | Resveratrol disulfates |
| RES | Resveratrol |
| TNFα | Tumor necrosis factor alpha |
References
- J. Burns, et al., Plant foods and herbal sources of resveratrol, J. Agric. Food Chem., 2002, 50(11), 3337–3340 CrossRef CAS PubMed.
- Y. Wang, et al., An LC-MS method for analyzing total resveratrol in grape juice, cranberry juice, and in wine, J. Agric. Food Chem., 2002, 50(3), 431–435 CrossRef CAS PubMed.
- S. He and X. Yan, From resveratrol to its derivatives: new sources of natural antioxidant, Curr. Med. Chem., 2013, 20(8), 1005–1017 CAS.
- K. van de Wetering, et al., Intestinal breast cancer resistance protein (BCRP)/Bcrp1 and multidrug resistance protein 3 (MRP3)/Mrp3 are involved in the pharmacokinetics of resveratrol, Mol. Pharmacol., 2009, 75(4), 876–885 CrossRef CAS PubMed.
- M. S. Baliga and S. K. Katiyar, Chemoprevention of photocarcinogenesis by selected dietary botanicals, Photochem. Photobiol. Sci., 2006, 5(2), 243–253 CAS.
- G. Bureau, F. Longpre and M. G. Martinoli, Resveratrol and quercetin, two natural polyphenols, reduce apoptotic neuronal cell death induced by neuroinflammation, J. Neurosci. Res., 2008, 86(2), 403–410 CrossRef CAS PubMed.
- G. Andersen, et al., High dose of dietary resveratrol enhances insulin sensitivity in healthy rats but does not lead to metabolite concentrations effective for SIRT1 expression, Mol. Nutr. Food Res., 2011, 55(8), 1197–1206 CAS.
- A. Burkon and V. Somoza, Quantification of free and protein-bound trans-resveratrol metabolites and identification of trans-resveratrol-C/O-conjugated diglucuronides - two novel resveratrol metabolites in human plasma, Mol. Nutr. Food Res., 2008, 52(5), 549–557 CAS.
- E. Wenzel, et al., Bioactivity and metabolism of trans-resveratrol orally administered to Wistar rats, Mol. Nutr. Food Res., 2005, 49(5), 482–494 CAS.
- D. J. Boocock, et al., Quantitation of trans-resveratrol and detection of its metabolites in human plasma and urine by high performance liquid chromatography, J. Chromatogr., B: Anal. Technol. Biomed. Life Sci., 2007, 848(2), 182–187 CrossRef CAS PubMed.
- D. J. Boocock, et al., Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent, Cancer Epidemiol., Biomarkers Prev., 2007, 16(6), 1246–1252 CAS.
- V. A. Brown, et al., Repeat dose study of the cancer chemopreventive agent resveratrol in healthy volunteers: safety, pharmacokinetics, and effect on the insulin-like growth factor axis, Cancer Res., 2010, 70(22), 9003–9011 CrossRef CAS PubMed.
- Y. Zong, et al., Resveratrol inhibits LPS-induced MAPKs activation via activation of the phosphatidylinositol 3-kinase pathway in murine RAW 264.7 macrophage cells, PLoS One, 2012, 7(8), e44107 CAS.
- R. R. Schumann, et al., Structure and function of lipopolysaccharide binding protein, Science, 1990, 249(4975), 1429–1431 CAS.
- S. Brandau, et al., Tissue-resident mesenchymal stem cells attract peripheral blood neutrophils and enhance their inflammatory activity in response to microbial challenge, J. Leukocyte Biol., 2010, 88(5), 1005–1015 CrossRef CAS PubMed.
- M. G. Rittig, et al., Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes, J. Leukocyte Biol., 2003, 74(6), 1045–1055 CrossRef CAS PubMed.
- J. Berthet, et al., Human platelets can discriminate between various bacterial LPS isoforms via TLR4 signaling and differential cytokine secretion, Clin. Immunol., 2012, 145(3), 189–200 CrossRef CAS PubMed.
- S. N. Verouti, et al., PAF effects on MCP-1 and IL-6 secretion in U-937 monocytes in comparison with oxLDL and IL-1beta effects, Atherosclerosis, 2011, 219(2), 519–525 CrossRef CAS PubMed.
- J. M. Walker, et al., Identification of Magnolia officinalis L. bark extract as the most potent anti-inflammatory of four plant extracts, Am. J. Chin. Med., 2013, 41(3), 531–544 CrossRef PubMed.
- C. H. Jang, et al., Chloroquine inhibits production of TNF-alpha, IL-1beta and IL-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes, Rheumatology, 2006, 45(6), 703–710 CrossRef CAS PubMed.
- V. Aires, et al., Resveratrol metabolites inhibit human metastatic colon cancer cells progression and synergize with chemotherapeutic drugs to induce cell death, Mol. Nutr. Food Res., 2013, 57(7), 1170–1181 CAS.
- J. Hoshino, et al., Selective synthesis and biological evaluation of sulfate-conjugated resveratrol metabolites, J. Med. Chem., 2010, 53(13), 5033–5043 CrossRef CAS PubMed.
- E. Wenzel and V. Somoza, Metabolism and bioavailability of trans-resveratrol, Mol. Nutr. Food Res., 2005, 49(5), 472–481 CAS.
- H. Yamamoto, et al., Sphingosylphosphorylcholine and lysosulfatide have inverse regulatory functions in monocytic cell differentiation into macrophages, Arch. Biochem. Biophys., 2011, 506(1), 83–91 CrossRef CAS PubMed.
- H. Inoue and T. Tanabe, Transcriptional role of the nuclear factor kappa B site in the induction by lipopolysaccharide and suppression by dexamethasone of cyclooxygenase-2 in U937 cells, Biochem. Biophys. Res. Commun., 1998, 244(1), 143–148 CrossRef CAS PubMed.
- F. H. Shand, et al., In vitro and in vivo evidence for anti-inflammatory properties of 2-methoxyestradiol, J. Pharmacol. Exp. Ther., 2011, 336(3), 962–972 CrossRef CAS PubMed.
- M. M. Ehrnhoefer-Ressler, et al., Identification of 1,8-cineole, borneol, camphor, and thujone as anti-inflammatory compounds in a Salvia officinalis L. infusion using human gingival fibroblasts, J. Agric. Food Chem., 2013, 61(14), 3451–3459 CrossRef CAS PubMed.
- J. Walker, et al., Identification of beer bitter acids regulating mechanisms of gastric acid secretion, J. Agric. Food Chem., 2012, 60(6), 1405–1412 CrossRef CAS PubMed.
- A. P. Piehler, et al., Gene expression results in lipopolysaccharide-stimulated monocytes depend significantly on the choice of reference genes, BMC Immunol., 2010, 11, 21 CrossRef PubMed.
- Y. Y. Lee, et al., Eugenol suppressed the expression of lipopolysaccharide-induced proinflammatory mediators in human macrophages, J. Endod., 2007, 33(6), 698–702 CrossRef PubMed.
- R. Lu and P. M. Pitha, Monocyte differentiation to macrophage requires interferon regulatory factor 7, J. Biol. Chem., 2001, 276(48), 45491–45496 CrossRef CAS PubMed.
- A. O'Hara, et al., Stimulation of inflammatory gene expression in human preadipocytes by macrophage-conditioned medium: upregulation of IL-6 production by macrophage-derived IL-1beta, Mol. Cell. Endocrinol., 2012, 349(2), 239–247 CrossRef PubMed.
- A. Bumrungpert, et al., Xanthones from mangosteen inhibit inflammation in human macrophages and in human adipocytes exposed to macrophage-conditioned media, J. Nutr., 2010, 140(4), 842–847 CrossRef CAS PubMed.
- E. Lupinacci, et al., Xanthohumol from hop (Humulus lupulus L.) is an efficient inhibitor of monocyte chemoattractant protein-1 and tumor necrosis factor-alpha release in LPS-stimulated RAW 264.7 mouse macrophages and U937 human monocytes, J. Agric. Food Chem., 2009, 57(16), 7274–7281 CrossRef CAS PubMed.
- T. Okoko and I. F. Oruambo, Inhibitory activity of quercetin and its metabolite on lipopolysaccharide-induced activation of macrophage U937 cells, Food Chem. Toxicol., 2009, 47(4), 809–812 CrossRef CAS PubMed.
- Y. T. Li, et al., Resveratrol inhibits matrix metalloproteinase-9 transcription in U937 cells, Acta Pharmacol. Sin., 2003, 24(11), 1167–1171 CAS.
- A. A. Qureshi, et al., Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by resveratrol, a potent proteasome inhibitor, Lipids Health Dis., 2012, 11, 76 CrossRef CAS PubMed.
- E. Derlindati, et al., Quercetin-3-O-glucuronide affects the gene expression profile of M1 and M2a human macrophages exhibiting anti-inflammatory effects, Food Funct., 2012, 3(11), 1144–1152 CAS.
- D. Delmas, et al., Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells, J. Biol. Chem., 2003, 278(42), 41482–41490 CrossRef CAS PubMed.
- J. Marcotorchino, et al., Lycopene attenuates LPS-induced TNF-alpha secretion in macrophages and inflammatory markers in adipocytes exposed to macrophage-conditioned media, Mol. Nutr. Food Res., 2012, 56(5), 725–732 CAS.
- H. Capiralla, et al., Resveratrol mitigates lipopolysaccharide- and Abeta-mediated microglial inflammation by inhibiting the TLR4/NF-kappaB/STAT signaling cascade, J. Neurochem., 2012, 120(3), 461–472 CrossRef CAS PubMed.
- E. Y. Chung, et al., Resveratrol down-regulates interferon-gamma-inducible inflammatory genes in macrophages: molecular mechanism via decreased STAT-1 activation, J. Nutr. Biochem., 2011, 22(10), 902–909 CrossRef CAS PubMed.
- C. C. Chan, et al., The protective role of natural phytoalexin resveratrol on inflammation, fibrosis and regeneration in cholestatic liver injury, Mol. Nutr. Food Res., 2011, 55(12), 1841–1849 CAS.
- Y. J. Jeon, et al., Dexamethasone inhibits IL-1 beta gene expression in LPS-stimulated RAW 264.7 cells by blocking NF-kappa B/Rel and AP-1 activation, Immunopharmacology, 2000, 48(2), 173–183 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |