 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bismuth(III) benzohydroxamates: powerful anti-bacterial activity against Helicobacter pylori and hydrolysis to a unique Bi34 oxido-cluster [Bi34O22(BHA)22(H-BHA)14(DMSO)6]†
Amita
Pathak
a,
Victoria L.
Blair
a,
Richard L.
Ferrero
b,
Michael
Mehring
c and
Philip C.
Andrews
*a
aSchool of Chemistry, Monash University, Clayton, Melbourne, VIC 3800, Australia. E-mail: phil.andrews@monash.edu
bMIMR-PHI Institute of Medical Research, Centre for Innate Immunity and Infectious Diseases Monash University, Clayton, Melbourne, VIC 3168, Australia
cFakultät für Naturwissenschaften, Institut für Chemie, Professur Koordinationschemie, Technische Universität Chemnitz, 09107 Chemnitz, Germany
First published on 17th October 2014
Abstract
Reaction of BiPh3 or Bi(OtBu)3 with benzohydroxamic acid (H2-BHA) results in formation of novel mono- and di-anionic hydroxamato complexes; [Bi2(BHA)3]∞1, [Bi(H-BHA)3] 2, [Bi(BHA)(H-BHA)] 3, all of which display nM activity against Helicobacter pylori. Subsequent dissolution of [Bi2(BHA)3]∞ in DMSO/toluene results in hydrolysis to the first structurally authenticated {Bi34} oxido-cluster [Bi34O22(BHA)22(H-BHA)14(DMSO)6] 4.
In conjunction with other antibiotics bismuth subsalicylate (BSS) and colloidal bismuth subcitrate (CBS) are taken therapeutically in the treatment and eradication of the ulcer and cancer causing bacterium Helicobacter pylori.1,2 Recent studies have begun to uncover mechanisms by which bismuth is toxic to the bacterium, including disrupting the enzymes urease and alcohol dehydrogenase, and interfering with Fe3+ regulating proteins.3 While having a strong affinity for S-based ligands, such as glutathione and metallothionein,4 Bi3+ is also known to compete with ferric ions in vivo with predominantly N- and O-binding biomolecules.5 In human serum this involves strong complexation with transferrin and lactoferrin, and it has been speculated that this is an important factor in the unusually low toxicity of bismuth in humans.6 In H. pylori, Fe3+ limitation mimics the antimicrobial effects of bismuth, though the mode of action appears unrelated, while increasing Fe3+ concentration acts to protect the bacterium against low concentrations of Bi3+ by outcompeting the heavy metal for transport proteins.7
To survive many bacteria rely on the ability to sequester and transport ferric ions by use of strong metal chelators, known as siderophores.8 These are predominantly polydentate O-binding molecules with carboxylate, catecholate and hydroxamate functional groups. Evidence suggests, however, that H. pylori does not generate siderophores but obtains iron from host transferrin and lactoferrin,9 and indeed H. pylori infection is associated with iron deficiency anaemia.10
While the chemistry of bismuth carboxylates has been relatively well studied,11,12 there have been no reports on the synthesis and structural chemistry of bismuth(III) complexes derived from hydroxamic acids. Carboxylates are known to form poorly soluble polymeric species, and can be hydrolytically unstable. As a result polynuclear oxido-clusters can form, as occurs in the formation of bismuth subsalicylate, [Bi38O44(HSal)26(Me2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)16(H2O)2],13 and bismuth subcitrate, (NH4)6[Bi6O4OH(cit)3(H2O)3](H2O)2,14 (NH4)12[Bi12O8(cit)8](H2O)10.15
O)16(H2O)2],13 and bismuth subcitrate, (NH4)6[Bi6O4OH(cit)3(H2O)3](H2O)2,14 (NH4)12[Bi12O8(cit)8](H2O)10.15
Antibiotic resistance, particularly to clarithromycin and metronidazole, is limiting the effectiveness of non-bismuth therapies for the treatment of H. pylori.16 Hydroxamic acids, particularly acetohydroxamic acid, are also known as urease inhibitors and so have been proposed as drug candidates for the treatment of H. pylori, though no compounds are yet clinically available.17 Taking this in conjunction with our knowledge of the close relationship of Bi3+ and Fe3+ in the lifecycle and killing of H. pylori, it is clear that bismuth(III) hydroxamates deserve closer examination as possible combination drugs for the treatment and eradication of H. pylori. In fact the role of hydroxamates as siderophores means they could have much broader application as novel anti-bacterial compounds.
In this paper we report the synthesis of the first bismuth(III) hydroxamate complexes: [Bi2(BHA)3] 1, [Bi(H-BHA)3] 2, [Bi(BHA)(H-BHA)] 3, as derived from benzohydroxamic acid (H2-BHA), and demonstrate its excellent in vitro activity against H. pylori. We also show that complex 1 undergoes slow hydrolysis in DMSO solution to give crystals of the unique Bi34 oxido-cluster [Bi34O22(BHA)22(H-BHA)14(DMSO)6] 4 (Scheme 1).
 | ||
| Scheme 1 Synthesis of bismuth hydroxamate complexes [Bi2(BHA)3]∞1, [Bi(H-BHA)3] 2, [Bi(BHA)(H-BHA)] 3, and the oxido-cluster [Bi34O22(BHA)22(H-BHA)14DMSO6] 4. | ||
The 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric reaction of H2-BHA with Bi(OtBu)3 in dry THF results in the clean generation of [Bi2(BHA)3] 1 in 66% yield. The low temperature addition of Bi(OtBu)3 prevents a rapid reduction of Bi(III) to Bi(0) as indicated by the appearance of a grey solid. Complex 1, comprises only hydroxamate di-anions. The solid was found to be only soluble in the highly polar solvents DMSO and DMF.
1 stoichiometric reaction of H2-BHA with Bi(OtBu)3 in dry THF results in the clean generation of [Bi2(BHA)3] 1 in 66% yield. The low temperature addition of Bi(OtBu)3 prevents a rapid reduction of Bi(III) to Bi(0) as indicated by the appearance of a grey solid. Complex 1, comprises only hydroxamate di-anions. The solid was found to be only soluble in the highly polar solvents DMSO and DMF.
In an attempt to obtain crystals of the complex the solid was dissolved in DMSO/toluene. However, rather than crystals of 1 the pale yellow crystals which formed are of a unique bismuth oxido-cluster; [Bi34O22(BHA)22(H-BHA)14DMSO6] 4. Formed slowly through hydrolysis this oxido-cluster contains both the H-BHA mono-anion, PhC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–N(H)–O−, as well as the expected BHA di-anion, PhC(–O−)
O)–N(H)–O−, as well as the expected BHA di-anion, PhC(–O−)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–O−.
N–O−.
Interestingly, the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric reaction of H2-BHA with Bi(OtBu)3 in dry THF resulted only in the isolation of the mixed anion species; [Bi(BHA)(H-BHA)] 3 in 75% yield (based on Bi). To obtain the fully substituted product [Bi(H-BHA)3], the weaker base BiPh3 was necessary. This allows for only a single deprotonation to give the H-BHA mono-anion. Complex 3 was formed by solvent-mediated reactions, in toluene (24 h, yield 58%) or ethanol (24 h, yield 55%) at reflux, but in higher yield under solvent free conditions (60 °C, 4 h, yield 65%).
1 stoichiometric reaction of H2-BHA with Bi(OtBu)3 in dry THF resulted only in the isolation of the mixed anion species; [Bi(BHA)(H-BHA)] 3 in 75% yield (based on Bi). To obtain the fully substituted product [Bi(H-BHA)3], the weaker base BiPh3 was necessary. This allows for only a single deprotonation to give the H-BHA mono-anion. Complex 3 was formed by solvent-mediated reactions, in toluene (24 h, yield 58%) or ethanol (24 h, yield 55%) at reflux, but in higher yield under solvent free conditions (60 °C, 4 h, yield 65%).
Full synthetic details and compound characterization for 1–4 are provided in the ESI.† NMR spectra were obtained on 1–4 in dry d6-DMSO. In the 1H NMR spectrum of [Bi2(BHA)3] 1, the resonances assigned to the NH and OH in benzohydroxamic acid, at 11.19 and 9.05 ppm respectively are absent (Fig. S5, ESI†). While the o-H and p-H protons shift to a lower frequency on complexation with Bi (7.43 to 7.33 and 7.50 to 7.33 ppm), the m-H protons shift to higher frequency (7.76 to 7.70 ppm). Suggesting electronic and structural fluxionality in the H-BHA ligand, the spectrum of 2 (Fig. S8, ESI†) shows resonances for both NH (0.5 H) and OH (0.5 H) at 11.14 and 9.02 ppm respectively. In contrast, complex 3 shows only a broad NH resonance at the higher frequency value of 12.48 ppm (Fig. S11, ESI†). The spectrum of the oxido-cluster 4 has the most acidic NH with the proton resonating at 13.66 ppm (Fig. S13, ESI†).
The sensitivity of complex 1 to hydrolysis is apparent in the 1H NMR spectrum collected in d6-DMSO, wherein the relative integral value for the NH signal at 13.60 ppm, when it becomes apparent, increases slowly over time. A similar behaviour of hydrolysis was recently reported for various bismuth oxido-nitrates and carboxylates, mainly on the basis of electrospray mass spectrometry and isolation of crystalline intermediates.18–20 So far, single crystal X-ray structure analyses exist for the most prominent cluster nuclearities {Bi6}, {Bi22} and {Bi38}. Cluster 4 represents a novel and unique intermediate of {Bi34} within this series.
Its molecular structure (Fig. 1) is composed of 34 bismuth atoms, 22 O2− anions, 22 di-anionic and 14 mono-anionic hydroxamate ligands, as well as 6 coordinated DMSO molecules. To the best of our knowledge this is the first structurally characterised Bi34-oxido cluster, with the heterometallic Bi/Na cluster [Bi33NaO38(OSiMe3)24] the closest comparison with regard to nuclearity.21
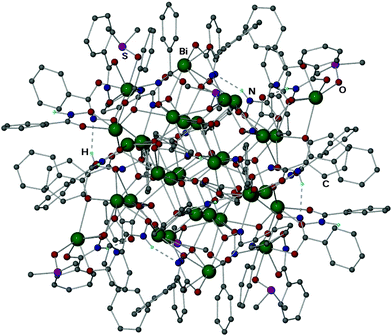 | ||
| Fig. 1 Molecular structure of [Bi34O22(BHA)22(H-BHA)14DMSO6] 4. Hydrogen atoms (except N–H) have been omitted for clarity. Selected bond lengths (Å) and angles (°) listed in the ESI.† Colour scheme: Bi, green; O, red; N, blue; S, purple; C, grey. | ||
The cluster has a central Bi24 unit [Bi(1)–Bi(12)], held together by 16 μ4-oxido and 6 μ3-oxido ligands, which can be easily distinguished with a Bi–O bond range of 2.098(10)–2.747(11) Å. The Bi24O22 core contains six edge sharing polyhedral units, of which two in the center, are constructed of [Bi6O7]4+ units (Fig. 2 red polyhedro) while the remaining four consist of [Bi6O6]6+ units (Fig. 2, blue polyhedro), all connected through their Bi–Bi edges. The two different {Bi6} units in 4 can be considered to be oxygen deficient and unique in its composition when compared to the more commonly seen hexameric unit [Bi6O8]2+ present in a wide variety of previously reported molecular Bi-oxido clusters.22 Making up the final Bi34 composition, the core of 4 is then encapsulated by a further 10 Bi atoms, each of which is bound to the Bi24-oxido core by either mono- or di-anionic HBA ligands, or a mixture of both.
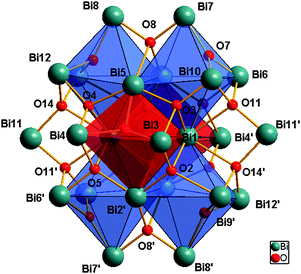 | ||
| Fig. 2 View of the [Bi24O22] central core in 2 with octahedral [Bi6O7]4+ and [Bi6O6]5+ units highlighted in red and blue respectively. | ||
The Bi24-oxido core of 4 can be envisaged as a Bi22-oxido core, having three bismuth containing layers, with two additionally attached bismuth atoms to the central layer. Both {Bi22} and {Bi24} each represent a cut-out of high-nuclearity {Bi38} oxido clusters, e.g. [Bi38O44(HSal)26(Me2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)16(H2O)2]13 and [Bi38O45(HSal)22(OH)2(DMSO)16.5].23 It might be assumed that clusters with such a {Bi24}-oxido core are representative of the hydrolysis step directly following the prominent {Bi22} clusters (Fig. S1–S3, ESI†).21 Additionally, two bismuth atoms (Bi14 and Bi17 in Fig. S4, ESI†) could be regarded as being on their way to attach to the cluster core upon additional hydrolysis. However, not all of the peripheral 10 bismuth atoms of 4 are found in positions close to those in the highly condensed {Bi38O45} clusters and it is assumed that dynamic coordination behaviour is crucial to further cluster growth processes. Most probably, coordination of single bismuth atoms (rather than larger moieties) prior to hydrolysis/condensation at the periphery of the cluster is an important step upon cluster growth, demonstrated here for the first time by a snapshot. It is assumed that ligands bound to the surface will control the kinetics of cluster growth, as it is reported for example for the growth of nanoparticles using surfactants.24
O)16(H2O)2]13 and [Bi38O45(HSal)22(OH)2(DMSO)16.5].23 It might be assumed that clusters with such a {Bi24}-oxido core are representative of the hydrolysis step directly following the prominent {Bi22} clusters (Fig. S1–S3, ESI†).21 Additionally, two bismuth atoms (Bi14 and Bi17 in Fig. S4, ESI†) could be regarded as being on their way to attach to the cluster core upon additional hydrolysis. However, not all of the peripheral 10 bismuth atoms of 4 are found in positions close to those in the highly condensed {Bi38O45} clusters and it is assumed that dynamic coordination behaviour is crucial to further cluster growth processes. Most probably, coordination of single bismuth atoms (rather than larger moieties) prior to hydrolysis/condensation at the periphery of the cluster is an important step upon cluster growth, demonstrated here for the first time by a snapshot. It is assumed that ligands bound to the surface will control the kinetics of cluster growth, as it is reported for example for the growth of nanoparticles using surfactants.24
The bactericidal activity of benzohydroxamic acid and complexes 1–4 was assessed against three laboratory strains of H. pylori: B128, 251 and 26695, using compound concentrations ranging from 25 to 0.024 μg mL−1. The Minimum Inhibitory Concentration (MIC) of each was determined by the Agar Diffusion method. The benzohydroxamic acid showed no activity against any of the H. pylori strains >25 μg mL−1. In contrast, as shown in Table 1, the bismuth complexes display excellent anti-bacterial activity against all three strains. Interestingly, any slow hydrolysis that takes the di-anion BHA (1) to the mono-anion H-BHA (2, 3) within a complex appears to have little or no impact on the bactericidal properties.
| Compound | H. pylori strain | ||
|---|---|---|---|
| 26695 | B128 | 251 | |
| Benzohydroxamic acid | >25 | >25 | >25 |
| [Bi2(BHA)3] 1 | 0.19 (0.23) | 0.19 (0.23) | 1.56 (1.89) |
| [Bi(H-BHA)3] 2 | 0.09 (0.14) | 0.05 (0.08) | 0.78 (1.26) |
| [Bi(BHA)(H-BHA)] 3 | 0.19 (0.39) | 0.09 (0.19) | 1.56 (3.24) |
| [Bi34O22(BHA)22(H-BHA)14(DMSO)6] 4 | 3.12 (0.24) | 6.25 (0.48) | 3.12 (0.24) |
These are remarkably low MIC values, with the only close comparison being with some amino-arenesulfonates which showed activities as low as MIC 0.05 μg mL−1 (0.06 μM), e.g. [Bi(O3S-(o-AB))3] from ortho-aminobenzenesulfonic acid ((o-AB)SO3H).25 As expected, the cluster is not so active, yet MIC values are all significantly lower than for the clinically available compounds: BSS and CBS, 12.5 μg mL−1; and RBC 8 μg mL−1.26 The bismuth benzohydroxamates are by far more effective than other Bi(III) complexes we have studied. For example, mono-nuclear carboxylates27 and thiobenzoates, [BiL3], generally gave MIC values of 6.25 μg mL−1,28 and thioxoketones 3.12 μg mL−1.29
These results indicate that both the structural chemistry and the anti-bacterial activity of bismuth(III) hydroxamates requires further investigation. As such, we are now currently expanding the breadth of complexes being studied.
Notes and references
- R. Ge, Z. Chen and Q. Zhou, Metallomics, 2012, 4, 239 RSC.
- J. P. Gisbert, World J. Gastroenterol., 2008, 14, 5385 CrossRef CAS PubMed.
- N. Yang and H. Sun, Coord. Chem. Rev., 2007, 251, 2354 CrossRef CAS.
- R. Ge and H. Sun, Acc. Chem. Res., 2007, 40, 267 CrossRef CAS PubMed.
- H. Li and H. Sun, Curr. Opin. Chem. Biol., 2012, 16, 74 CrossRef CAS PubMed.
- H. Sun and K. Y. Szeto, J. Inorg. Biochem., 2003, 94, 114 CrossRef CAS PubMed.
- R. Ge and H. Sun, Biometals, 2012, 25, 247 CrossRef CAS PubMed.
- J. B. Nielands, J. Biol. Chem., 1995, 270, 26723 CrossRef.
- O. Senkovich, S. Ceaser, D. J. McGee and T. L. Testerman, Infect. Immun., 2010, 78, 1841 CrossRef CAS PubMed.
- P. Malfertheiner and M. Selgrad, Curr. Opin. Gastroenterol., 2010, 26, 618 CrossRef PubMed.
- P. C. Andrews, G. B. Deacon, P. C. Junk, I. Kumar and M. Silberstein, Dalton Trans., 2006, 4852 RSC.
- V. Stavila, R. L. Davidovich, A. Gulea and K. H. Whitmire, Coord. Chem. Rev., 2006, 250, 2782 CrossRef CAS.
- P. C. Andrews, G. B. Deacon, P. C. Junk and M. Maguire, Angew. Chem., Int. Ed., 2006, 45, 5638 CrossRef CAS PubMed.
- E. Asato, K. Katsura, M. Mikuriya, T. Fujii and J. Reedijk, Chem. Lett., 1992, 1967 CrossRef CAS.
- E. Asato, K. Katsura, M. Mikuriya, U. Turpeinen, I. Mutikainen and J. Reedijk, Inorg. Chem., 1995, 34, 2447 CrossRef CAS.
- F. Mégraud, Gut, 2004, 53, 1374 CrossRef PubMed.
- E. M. F. Muri and T. G. Barros, in Hydroxamic Acids, ed. S. P. Gupta, Pub. Springer, 2013, ch. 9 Search PubMed.
- M. Schlesinger, A. Pathak, S. Richter, D. Sattler, A. Seifert, T. Rüffer, P. C. Andrews, C. A. Schalley, H. Lang and M. Mehring, Eur. J. Inorg. Chem., 2014, 4218 CrossRef CAS.
- L. Miersch, M. Schlesinger, R. W. Troff, C. A. Schalley, T. Rüffer, H. Lang, D. Zahn and M. Mehring, Chem. – Eur. J., 2011, 17, 6985 CrossRef CAS PubMed.
- D. Sattler, M. Schlesinger, M. Mehring and C. A. Schalley, ChemPlusChem, 2013, 78, 1005 CrossRef CAS.
- M. Mehring, D. Mansfeld, S. Paalasmaa and M. Schürmann, Chem. – Eur. J., 2006, 12, 1767 CrossRef CAS PubMed.
- M. Mehring, Coord. Chem. Rev., 2007, 251, 974 CrossRef CAS.
- D. Mansfeld, L. Miersch, T. Rüffer, D. Schaarschmidt, H. Lang, T. Böhle, R. W. Troff, C. A. Schalley, J. Müller and M. Mehring, Chem. – Eur. J., 2011, 17, 14805 CrossRef CAS PubMed.
- Y. Yin and A. P. Alivisatos, Nature, 2005, 437, 664 CrossRef CAS PubMed.
- M. Busse, I. Trinh, P. C. Junk, R. L. Ferrero and P. C. Andrews, Chem. – Eur. J., 2013, 19, 5264 CrossRef CAS PubMed.
- (a) C. E. Haas, D. E. Nix and J. J. Schentag, Antimicrob. Agents Chemother., 1990, 34, 1637 CrossRef CAS PubMed; (b) Y. Glupczynski, M. Delmee, C. Bruck, M. Labbe, V. Avesani and A. Burette, Eur. J. Epidemiol., 1988, 4, 154 CrossRef CAS PubMed; (c) C. A. McNulty, J. Dent and R. Wise, Antimicrob. Agents Chemother., 1985, 28, 837 CrossRef CAS PubMed.
- P. C. Andrews, R. L. Ferrero, P. C. Junk, I. Kumar, Q. Luu, K. Nguyen and J. W. Taylor, Dalton Trans., 2010, 39, 2861 RSC.
- P. C. Andrews, R. L. Ferrero, P. C. Junk, J. G. MacLellan and R. M. Peiris, Aust. J. Chem., 2012, 65, 883 CrossRef CAS.
- P. C. Andrews, V. L. Blair, R. L. Ferrero, P. C. Junk, L. Kedzierski and R. M. Peiris, Dalton Trans., 2014, 43, 1279 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Full experimental and analytical details, including crystallographic data and tables. CCDC 1023760. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4cc07329k |
| This journal is © The Royal Society of Chemistry 2014 |
