Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
QM/MM simulations as an assay for carbapenemase activity in class A β-lactamases†
Ewa I.
Chudyk
a,
Michael A. L.
Limb
a,
Charlotte
Jones
a,
James
Spencer
b,
Marc W.
van der Kamp
 *a and
Adrian J.
Mulholland
*a
*a and
Adrian J.
Mulholland
*a
aCentre for Computational Chemistry, School of Chemistry, University of Bristol, Cantock's Close, Bristol, BS8 1TS, UK. E-mail: marc.vanderkamp@bristol.ac.uk; adrian.mulholland@bristol.ac.uk
bSchool of Cellular and Molecular Medicine, University of Bristol, University Walk, Bristol, BS8 1TD, UK
First published on 2nd October 2014
Abstract
Carbapenems, ‘last resort’ antibiotics for many bacterial infections, can now be broken down by several class A β-lactamases (i.e. carbapenemases). Here, carbapenemase activity is predicted through QM/MM dynamics simulations of acyl–enzyme deacylation, requiring only the 3D structure of the apo-enzyme. This may assist in anticipating resistance and future antibiotic design.
Antibiotic (antimicrobial) resistance, particularly in Gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa,1 has emerged as an extremely serious and growing medical problem. It threatens modern medicine, not only due to the growing difficulty of treating bacterial infections, but also jeopardizing many therapies and surgical procedures.2 Much of the developing resistance in such bacteria can be attributed to β-lactamases, enzymes which catalyse the breakdown of the essential β-lactam ring present in all classes of β-lactam antibiotics.3 Carbapenems are a highly potent class of antibiotics: they have a broad spectrum of antibacterial activity and, in contrast to many other classes of antibiotic, have not been susceptible to resistance due to hydrolysis by β-lactamases. The characteristic broad-spectrum antibacterial activity exhibited by carbapenems means that they are currently used as effective treatments for otherwise incurable bacterial infections.4
Alarmingly, however, a variety of carbapenem-hydrolysing β-lactamases have been reported5 and are becoming increasingly common. These β-lactamases, known as carbapenemases, are able to efficiently hydrolyse the β-lactam ring in carbapenems, rendering this formerly highly potent class of antibiotics inactive and resulting in the re-emergence of potentially untreatable bacterial infections.6 The most common enzymes with carbapenemase activity are β-lactamases of class A (exemplified by the SFC-1 and KPC-2 enzymes7) and class D. Class B β-lactamases, such as NDM-1, are also increasingly prevalent carbapenemases.8 Here, we focus on β-lactamases of class A due to the growing frequency with which carbapenem-hydrolysing enzymes are encountered in both clinical and environmental bacteria. Carbapenemase activity in these enzymes is due to efficient deacylation; the majority of class A enzymes are inhibited by carbapenems because a long-lived acyl–enzyme complex is formed.
The first step of the class A β-lactamase hydrolysis mechanism is formation of an acyl–enzyme intermediate (via nucleophilic attack by an active site serine), with coincident opening of the β-lactam ring.9 Deacylation is then required to release the cleaved β-lactam and allow enzyme turnover. Deacylation is initiated by nucleophilic attack of a conserved water molecule (the deacylating water molecule, DW) on the ester carbon of the acyl–enzyme (Fig. 1). The water molecule is activated by proton abstraction by a glutamate residue situated in the active site acting as a base.10
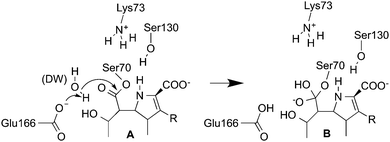 | ||
| Fig. 1 The first step of carbapenem deacylation: the acyl–enzyme (A) reacts to form a tetrahedral intermediate (B) (numbering for the TEM-1 enzyme).10 | ||
Carbapenem-inhibited class A β-lactamases are acylated readily but have a greatly reduced rate of deacylation,11 resulting in a long-lived acyl–enzyme. In contrast, β-lactamases with carbapenemase activity deacylate efficiently (with a short-lived acyl–enzyme), conferring carbapenem resistance on bacteria that carry these enzymes. Despite multiple structural12,13 and biochemical studies14,15 on class A β-lactamases, the difference in carbapenemase activity exhibited by different β-lactamases (which have highly homologous active sites) is not well understood. Recently, we suggested that the orientation of the 6α-1R-hydroxyethyl group of carbapenems in the enzyme active site may have an important influence on the deacylation rate (based on crystal structures and molecular dynamics (MD) simulations of the carbapenemase SFC-1).7 We proposed that, in carbapenemases, a conformation of the 6α-1R-hydroxyethyl that does not interfere with the nucleophilic attack of the DW is favored, which facilitates deacylation. Further subtle structural differences were identified that may affect the rate of deacylation, supporting previous findings.14,16,17 Here we use simulations to investigate class A β-lactamases, with the aim of identifying determinants of carbapenemase activity, and ultimately of applying these principles to assess the ability of uncharacterized enzymes to hydrolyse carbapenems. The need to recognize new and emerging resistance threats is growing in importance as new enzymes continue to be identified and new sequences emerge from large-scale sequencing projects.
We employ hybrid quantum mechanics/molecular mechanics (QM/MM) umbrella sampling MD calculations to investigate carbapenem hydrolysis in class A β-lactamases.18 Because the difference between carbapenemases and carbapenem-inhibited enzymes is in the deacylation step, and tetrahedral intermediate (TI) formation is likely to have the highest barrier in this reaction,11 we model the first step of deacylation only (Fig. 1). Simulations were performed with the semi-empirical SCC-DFTB QM method19 in the AMBER12 simulation package,20 using the ff12SB MM force-field for the protein, the TIP4P-Ew water model and the General AMBER Force Field (GAFF) for the part of meropenem not in the QM region. The QM region (Fig. S1, ESI†) consisted of the common carbapenem scaffold (omitting the 3-[5-(dimethylcarbamoyl) pyrrolidin-2-y]-sulfanyl R-group when modelling meropenem, see Fig. 1), DW and the Ser70 and Glu166 sidechains from Cβ. The protocol can be summarized as follows (see ESI† for details):
(1) Generate and solvate the acyl–enzyme complex (AC) starting structure. In absence of an acyl–enzyme crystal structure, dock meropenem into the protein in the conformation observed crystallographically in the SFC-1 acyl–enzyme (PDB ID: 4EV4).
(2) Heat the system to 300 K and equilibrate at 300 K and 1 atm in QM/MM MD (see ESI†).
(3) 300 ps of unrestrained QM/MM MD of the acyl–enzyme.
(4) Select a starting structure for reaction modelling. Previous work7 indicated that three distinct conformations of the 6α-1R-hydroxyethyl group could be present in the acyl–enzyme complex. A suitable structure for reaction modelling must satisfy two criteria: (i) it contains the predominant conformation of the 6α-1R-hydroxyethyl observed over the course of 300 ps unrestrained MD simulation and (ii) it contains the DW correctly positioned for nucleophilic attack of the electrophilic carbon in the ligand (indicated by a distance of less than 3.5 Å). If both cannot be satisfied, steps 1–3 are repeated with the 6α-1R-hydroxyethyl conformation as in the structure with PDB ID: 4EUZ.7
(5) Perform QM/MM umbrella sampling MD along two reaction coordinates: rx = d(Oε1Glu166–H2DW) − d(H2DW–ODW) (proton transfer from DW to Glu166) and ry = d(CCMer–ODW) (nucleophilic attack of DW on the carbonyl carbon) to simulate the deacylation reaction. Perform 20 ps of MD for each simulation window. We repeated this step three times to test convergence.
(6) Calculate the 2D free energy surface for the reaction using the weighted-histogram analysis method (WHAM) and identify the minimum energy path (MEP) on this surface; the highest point along the MEP is taken as the transition state, giving the activation free energy, ΔG‡calc (Fig. 2).
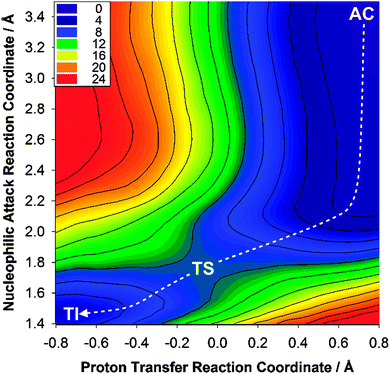 | ||
| Fig. 2 Free energy surface for the deacylation of meropenem in KPC-2. AC: acyl–enzyme complex; TS: transition state; TI: tetrahedral intermediate. | ||
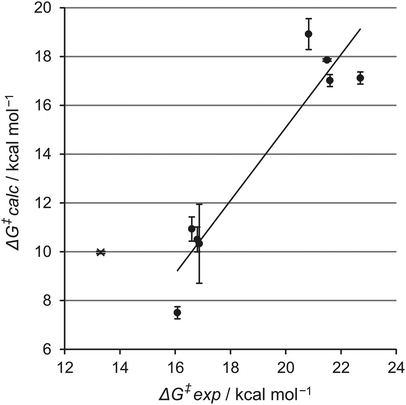
The carbapenemase activity of eight different class A β-lactamases was assessed using this simulation assay: these were the carbapenemases KPC-2, NMC-A, SFC-1 and SME-1 and the carbapenem-inhibited β-lactamases CTX-M-16, BlaC, TEM-1, and SHV-1. Additionally, the deacylation of benzylpenicillin in TEM-1 was simulated for comparison and to investigate the generality of the method with different β-lactam antibiotics. Crystallographic carbapenem acyl–enzyme structures of wild-type TEM-1, SHV-1 and BlaC and a SFC-1 mutant (E166A), and TEM-1 with benzylpenicillin were available. For KPC-2, SME-1, NMC-A and CTX-M-16, the crystal structure of the apo-form was used as the starting point (see Table S1, ESI†). For all eight enzymes, experimental kinetic data are available, enabling comparison of the calculated barriers with experiment (Table 1).
| β-Lactamase | k cat (s−1) | ΔG‡exp (kcal mol−1) | ΔG‡calcb (kcal mol−1) |
|---|---|---|---|
| a Calculation performed with benzylpenicillin. b Calculated barriers are the average from three separate simulations; standard deviation in parenthesis. | |||
| BlaC21 | 1.7 × 10−3 | 21.5 | 17.9 (0.08) |
| CTX-M22 | 4.2 × 10−3 | 20.8 | 18.9 (1.09) |
| SHV-122 | 1.3 × 10−3 | 21.6 | 17.0 (0.43) |
| TEM-122 | 2.3 × 10−3 | 22.7 | 17.1 (0.43) |
| KPC-222 | 3.6 | 16.8 | 10.5 (0.88) |
| NMC-A23 | 12.0 | 16.1 | 7.5 (0.43) |
| SFC-124 | 6.5 | 16.6 | 10.9 (0.86) |
| SME-122 | 3.2 | 16.9 | 10.3 (2.80) |
TEM-1a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 22 |
1500 | 13.3 | 10.0 (0.04) |
The results indicate that the protocol demonstrated here correctly distinguishes carbapenemases from carbapenem-inhibited class A β-lactamases. A low calculated activation free energy of ∼10 kcal mol−1 signifies efficient deacylation (and therefore carbapenemase activity) whereas higher calculated activation free energies of 17–18 kcal mol−1 show a long-lived acyl–enzyme (and therefore a carbapenem-inhibited enzyme). This is true both for simulations run from experimentally determined acyl–enzyme structures and for those with only a crystal structure of the apo enzyme as input.
In both cases, comparison with experimental data (Table 1; Fig. 3) reveals a clear discrimination between the two types of enzyme. Moreover, application to the reaction of TEM-1 with benzylpenicillin indicates that the same protocol can also predict activity against other classes of antibiotics.
Comparison of the experimental and calculated barriers indicates that the SCC-DFTB/AMBER12SB QM/MM method underestimates the free activation energy, by 6.6 kcal mol−1 on average for carbapenemases, and 4.1 kcal mol−1 on average for the carbapenem-inhibited enzymes. This is due to the SCC-DFTB method, which is known to underestimate barriers in many cases.19,25 The QM/MM MD simulations with SCC-DFTB clearly distinguish between the two enzyme groups. They are computationally affordable, in contrast to equivalent simulations with DFT or ab initio methods for the QM region (these methods are more suitable for e.g. establishing reaction mechanisms9,10). For the purpose of screening enzyme activity relatively rapidly, semi-empirical methods (validated for the reaction of interest) thus remain highly attractive due to their low computational cost.
A vitally important question is understanding the molecular basis for carbapenemase activity in class A β-lactamases. Common structural and mechanistic features that differentiate carbapenemases from carbapenem-inhibited enzymes have not been identified. Many features, including active site volume13 and mobility, the propensity of the carbapenem acyl–enzyme to tautomerise into the less active Δ1-pyrroline form,26 the orientation of the carbapenem 6α-1R-hydroxymethyl group7,27 and the hydrogen bonding pattern of the deacylating water molecule,16 have been suggested to contribute to efficient carbapenem hydrolysis. Our simulations show no obvious common feature determining carbapenemase activity. Instead, multiple subtle factors appear to be at play, stressing the need for detailed investigations of individual reactions. Consistent with the conclusion that multiple subtle effects discriminate class A beta-lactamases that are carbapenemases from those that are not, inspection of our simulations suggests different effects in the different enzymes. For example, we have previously suggested that one important factor could be the orientation of the 6α-1R-hydroxymethyl group: when the hydroxyl forms a hydrogen bond with the deacylating water, the nucleophilic attack of this water on the acyl–enzyme carbonyl is hampered.7 Although this interaction is indeed observed in the simulations of one of the carbapenem-inhibited enzymes (TEM-1), it is not for the others. Additional simulations and analysis will be required to dissect the subtle differences in interactions that define the catalytic properties of the different individual enzymes. The data presented here demonstrate that the first-principles QM/MM reaction simulations performed here are able to incorporate these multiple relevant subtle effects, as demonstrated by the ability to predict the deacylation activity of the multiple systems under investigation. Indeed, detailed investigation of reactivity (and not e.g. simply studies of acyl–enzyme dynamics) will probably be necessary to understand the origins of carbapenemase activity.
In conclusion, the results of our reaction simulations of eight different class A β-lactamases show that QM/MM activation free energies for formation of the TI in carbapenem deacylation, obtained using a standard protocol involving semi-empirical QM/MM MD simulations, correctly predict their carbapenemase activity in silico. The QM/MM protocol presented here thus provides a computational assay to predict carbapenemase activity, based on only the 3D structure of class A β-lactamases. QM/MM simulations can thus play a vital role in the assessment and understanding of emerging β-lactam resistance and are potentially useful in the development of new antibiotics.
We thank the U.K. Engineering and Physical Science Research Council (EPSRC; grant no. EP/G007705/1) for support. MALL thanks EPSRC & SCI. JS thanks the U.K. Medical Research Council (MRC; U.K.-Canada Team Grant G1100135) and the U.S. National Institutes of Health (1R01AI100560-01). This work was conducted using the computational facilities of the Advanced Computing Research Centre, University of Bristol.
Notes and references
- D. M. Livermore, Korean J. Intern. Med., 2012, 27, 128–142 CrossRef CAS PubMed.
- World Health Organization, Antimicrobial resistance: global report on surveillance, 2014.
- J. F. Fisher, S. O. Meroueh and S. Mobashery, Chem. Rev., 2005, 105, 395–424 CrossRef CAS PubMed.
- D. P. Nicolau, Expert Opin. Pharmacother., 2008, 9, 23–37 CrossRef CAS PubMed; K. M. Papp-Wallace, A. Endimiani, M. A. Taracila and R. A. Bonomo, Antimicrob. Agents Chemother., 2011, 55, 4943–4960 CrossRef PubMed.
- H. Yigit, A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush and F. C. Tenover, Antimicrob. Agents Chemother., 2001, 45, 1151–1161 CrossRef CAS PubMed; I. Henriques, A. Moura, A. Alves, M. J. Saavedra and A. Correia, Antimicrob. Agents Chemother., 2004, 48, 2321–2323 CrossRef PubMed.
- P. Nordmann, G. Cuzon and T. Naas, Lancet Infect. Dis., 2009, 9, 228–236 CrossRef CAS.
- F. Fonseca, E. I. Chudyk, M. W. van der Kamp, A. Correia, A. J. Mulholland and J. Spencer, J. Am. Chem. Soc., 2012, 134, 18275–18285 CrossRef CAS PubMed.
- T. R. Walsh, Int. J. Antimicrob. Agents, 2010, 36(suppl 3), S8–S14 CrossRef CAS.
- J. C. Hermann, J. Pradon, J. N. Harvey and A. J. Mulholland, J. Phys. Chem. A, 2009, 113, 11984–11994 CrossRef CAS PubMed.
- J. C. Hermann, L. Ridder, H.-D. Höltje and A. J. Mulholland, Org. Biomol. Chem., 2006, 4, 206–210 CAS.
- L. Mourey, K. Miyashita, P. Swarén, A. Bulychev, J. P. Samama and S. Mobashery, J. Am. Chem. Soc., 1998, 120, 9382–9383 CrossRef CAS.
- P. Swarén, L. Maveyraud, X. Raquet, S. Cabantous, C. Duez, J. D. Pédelacq, S. Mariotte-Boyer, L. Mourey, R. Labia, M. H. Nicolas-Chanoine, P. Nordmann, J. M. Frère and J. P. Samama, J. Biol. Chem., 1998, 273, 26714–26721 CrossRef CAS PubMed; W. Sougakoff, G. L'Hermite, L. Pernot, T. Naas, V. Guillet, P. Nordmann, V. Jarlier and J. Delettre, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 267–274 CrossRef PubMed; S. Petrella, N. Ziental-Gelus, C. Mayer, M. Renard, V. Jarlier and W. Sougakoff, Antimicrob. Agents Chemother., 2008, 52, 3725–3736 CrossRef PubMed.
- W. Ke, C. R. Bethel, J. M. Thomson, R. A. Bonomo and F. van den Akker, Biochemistry, 2007, 46, 5732–5740 CrossRef CAS PubMed.
- G. Zafaralla and S. Mobashery, J. Am. Chem. Soc., 1992, 114, 1505–1506 CrossRef CAS.
- H. Frase, Q. Shi, S. A. Testero, S. Mobashery and S. B. Vakulenko, J. Biol. Chem., 2009, 284, 29509–29513 CrossRef CAS PubMed; F. K. Majiduddin and T. Palzkill, Antimicrob. Agents Chemother., 2003, 47, 1062–1067 CrossRef; F. K. Majiduddin and T. Palzkill, Antimicrob. Agents Chemother., 2005, 49, 3421–3427 CrossRef PubMed; K. M. Papp-Wallace, M. Taracila, J. M. Hornick, A. M. Hujer, K. M. Hujer, A. M. Distler, A. Endimiani and R. A. Bonomo, Antimicrob. Agents Chemother., 2010, 54, 2867–2877 CrossRef PubMed; K. M. Papp-Wallace, M. Taracila, C. J. Wallace, K. M. Hujer, C. R. Bethel, J. M. Hornick and R. A. Bonomo, Protein Sci., 2010, 19, 1714–1727 CrossRef PubMed.
- M. Nukaga, C. R. Bethel, J. M. Thomson, A. M. Hujer, A. Distler, V. E. Anderson, J. R. Knox and R. A. Bonomo, J. Am. Chem. Soc., 2008, 130, 12656–12662 CrossRef CAS PubMed.
- L. Maveyraud, L. Mourey, L. P. Kotra, J. D. Pédelacq, V. Guillet, S. Mobashery and J. P. Samama, J. Am. Chem. Soc., 1998, 120, 9748–9752 CrossRef CAS; C. R. Bethel, M. Taracila, T. Shyr, J. M. Thomson, A. M. Distler, K. M. Hujer, A. M. Hujer, A. Endimiani, K. Papp-Wallace, R. Bonnet and R. A. Bonomo, Antimicrob. Agents Chemother., 2011, 55, 3465–3475 CrossRef PubMed.
- M. W. van der Kamp and A. J. Mulholland, Biochemistry, 2013, 52, 2708–2728 CrossRef CAS PubMed.
- M. Elstner, T. Frauenheim, E. Kaxiras, G. Seifert and S. Suhai, Phys. Status Solidi B, 2000, 217, 357–376 CrossRef CAS; M. Gaus, Q. Cui and M. Elstner, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2014, 4, 49–61 CrossRef.
- D. A. Case, T. A. Darden, T. E. Cheatham III, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, R. C. Walker, W. Zhang, K. M. Merz, B. Roberts, S. Hayik, A. Roitberg, G. Seabra, J. Swails, A. W. Götz, I. Kolossváry, K. F. Wong, F. Paesani, J. Vanicek, R. M. Wolf, J. Liu, X. Wu, S. R. Brozell, T. Steinbrecher, H. Gohlke, Q. Cai, X. Ye, J. Wang, M.-J. Hsieh, G. Cui, D. R. Roe, D. H. Mathews, M. G. Seetin, R. Salomon-Ferrer, C. Sagui, V. Babin, T. Luchko, S. Gusarov, A. Kovalenko and P. A. Kollman, AMBER 12, University of California, San Francisco, 2012 Search PubMed.
- J.-E. Hugonnet, L. W. Tremblay, H. I. Boshoff, C. E. Barry III and J. S. Blanchard, Science, 2009, 323, 1215–1218 CrossRef CAS PubMed.
- A. M. Queenan, W. Shang, R. Flamm and K. Bush, Antimicrob. Agents Chemother., 2010, 54, 565–569 CrossRef CAS PubMed.
- M. S. Boyer, N. M. H. Chanoine and R. Labia, FEMS Microbiol. Lett., 1996, 143, 29–33 Search PubMed.
- F. Fonseca, A. C. Sarmento, I. Henriques, B. Samyn, J. van Beeumen, P. Domingues, M. R. Domingues, M. J. Saavedra and A. Correia, Antimicrob. Agents Chemother., 2007, 51, 4512–4514 CrossRef CAS PubMed.
- H. L. Woodcock, M. Hodoscek and B. R. Brooks, J. Phys. Chem. A, 2007, 111, 5720–5728 CrossRef CAS PubMed.
- M. Kelp and P. R. Carey, Biochemistry, 2008, 47, 11830–11837 CrossRef PubMed.
- C. A. Smith, H. Frase, M. Toth, M. Kumarasiri, K. Wiafe, J. Munoz, S. Mobashery and S. B. Vakulenko, J. Am. Chem. Soc., 2012, 134, 19512–19515 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Full description of methods, including PDB accession codes for all structures used (Table S1). Free energy surfaces for all reactions simulated (Fig. S2–S10). See DOI: 10.1039/c4cc06495j |
| This journal is © The Royal Society of Chemistry 2014 |