 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A robust DNA interface on a silicon electrode†
Pauline
Michaels
a,
Muhammad Tanzirul
Alam
a,
Simone
Ciampi
a,
William
Rouesnel
a,
Stephen G.
Parker
ab,
Moinul H.
Choudhury
a and
J. Justin
Gooding
*abc
aSchool of Chemistry, The University of New South Wales, Sydney, NSW 2052, Australia. E-mail: justin.gooding@unsw.edu.au; Fax: +61 2 9385 6141; Tel: +61 2 9385 5384
bAustralian Centre for NanoMedicine, The University of New South Wales, Sydney, NSW 2052, Australia
cARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW 2052, Australia
First published on 29th May 2014
Abstract
Two different interfaces prepared via UV-hydrosilylation of undecylenic acid and 1,8-nonadiyne on silicon(111) have been explored to develop a robust electrochemical DNA sensor. Electrodes modified with undecylenic acid were found to stably immobilise DNA but could not resist the growth of insulating oxides, whereas 1,8-nonadiyne modified electrodes satisfy both requirements.
Deoxyribonucleic acid (DNA) sequence detection has provided revolutionary benefits to a variety of fields, notably in pharmaceuticals, forensics and clinical diagnostics.1,2 The technique most frequently used to detect DNA sequencing is southern blotting,2 while electrochemical DNA biosensors have the potential of replacing this traditional labour intensive and time-consuming practice. Electrochemical DNA sensors also have the added advantage that the devices can be made portable. The effectiveness of DNA recognition interfaces for biosensing is dependent on the formation of highly stable DNA interfaces. Operations that might be conducted on such chips could include thermal cycling for polymerase chain reaction, repeated hybridization, and denaturation including elevated temperature or post hybridization modification of captured DNA.1,2 All these operations could challenge the integrity of a DNA interface.
DNA recognition interfaces are required to control the interaction between a surface bound DNA strand and a nucleic acid sequence in solution at the molecular level. As the result is a DNA hybrid, the restriction of the configuration freedom of the surface bound DNA needs to be minimised as much as possible. The most common strategies for doing this are via end point immobilisation of the DNA within, or onto, a self-assembled monolayer (SAM).1,2 The monolayer system most frequently used is of gold–thiol chemistry, but this surface chemistry has some stability issues with regards to limited potential window, long incubation times, influence of UV photooxidation, and elevated temperatures.3 A number of possible alternatives include the use of aryl diazonium salts,4 silane chemistry on oxide surfaces,5 and the hydrosilylation of alkenes at hydrogen terminated silicon.6 Aryl diazonium salts can be used to get enhanced stability, but the growth of layers on electrode surfaces is less controlled as compared to alkanethiol system. As a consequence surface become inhomogeneous and the film can be of multilayers7 with a thickness up to 10 nm.8 Surface chemistry of silanes9 has also been explored for the construction of sensing interfaces but these have compromised stability in aqueous media.10 Electroactive monolayers on silicon have been prepared by grafting different ferrocene compounds directly onto H-terminated silicon.11 Even though such reports describe well-behaved electronic communication between redox species and silicon, simultaneous protection against oxidation of the underlying silicon surface forming insulating oxide layer is either lacking or has not been investigated.
Hence, the application of silicon in electrochemistry requires effective passivation of the electrode such that the surface is essentially inaccessible to species in a solution. Organic monolayers bound directly to non-oxidized crystalline silicon surfaces through a silicon–carbon bond produce highly stable (80–100 kcal mol−1)12 and densely packed monolayers in comparison to conventional silane films on oxidized silicon. Such monolayers are easily used as platforms for further derivatization and have therefore become an advantageous approach in the fabrication of complex molecular architectures on surfaces.13 A further advantage is that it offers a myriad of detection options for DNA hybridization ranging from forming field effect transistors, electrochemical impedance, potentiometry and possibly even amperometry.
The purpose of this paper is to explore the relative merits of two strategies for immobilising DNA on silicon surfaces and monitoring DNA hybridization using Faradaic impedance spectroscopy. The passivating layers to inhibit the formation of insulating silicon oxides were formed via the UV-hydrosilylation14 of undecylenic acid or 1,8-nonadiyne on Si(111). The surface chemistries were evaluated from the perspective of getting selective DNA hybridization signals.
The detail procedures for fabricating the sensing interfaces (Scheme 1) are described in the ESI.† In order to gain information about the quality of the grafted monolayer and to differentiate between different target DNA analytes, modified surfaces (SAM-3) were characterised using Faradaic impedance spectroscopy. In this regard [Fe(CN)6]3−/4− was used as a redox probe. Upon hybridisation with complementary DNA, almost a two fold increase is observed in charge transfer resistance (Rct) as compared to the ss-DNA (%ΔRct = 192.0 ± 27%) at the undecylenic acid modified interface (see ESI†). This increase in Rct is comparable with that reported on gold surfaces.15 [Fe(CN)6]3−/4− is a highly charged anionic redox species and is thus repelled by the polyanionic DNA phosphate backbone. Rct increases as the negative charge on the electrode surface increases due to DNA hybridisation. An increase in Rct is also observed after exposure of ss-DNA to non-complementary DNA (%ΔRct = 75.4 ± 18%). In fact, with consecutive scans Rct continues to increase eventually equalling that of pure silicon oxides (11.8 MΩ). XPS spectra (see ESI†) at 102–105 eV confirms the formation of silicon oxides with potential cycling, while no peak is observed in a freshly prepared undecylenic acid modified silicon wafer. Allongue et al.16 correlate the oxide growth with pinhole defects in a grafted monolayer. The pinholes are most likely a result of electrostatic repulsion among the carboxylic acid moieties of the undecylenic acid.17 These defects act as an anchor point to the formation of oxide, which grows laterally, forming a monolayer underneath the immobilised organic layer. In addition, the immobilisation of DNA can also increase the corrosion (oxidation–hydrolysis) process of the silicon.18 Very little loss in the atomic percentage of carbon (22.1 and 17.5%) is observed before and after electrochemistry (i.e., with and without oxides), respectively, indicating that the undecylenic acid is still present on the surface. Although undecylenic acid modified surface has good stability with regards to immobilizing probe DNA, the surface chemistry fails to stabilise the silicon electronically. Hence, a surface chemistry that can satisfy both requirements is still required.
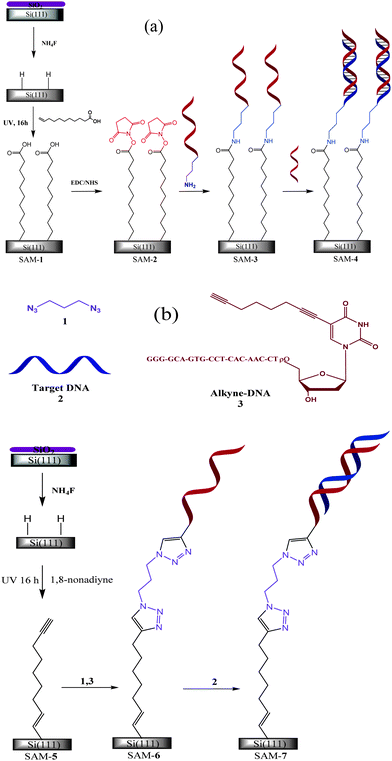 | ||
| Scheme 1 Two strategies (a) and (b) of immobilising DNA on Si(111) surface. In the first strategy surface was passivated by the UV-hydrosilylation of undecylenic acid and –NH2 terminated single stranded DNA (ss-DNA) was immobilised by forming amide bond with –COOH of undecylenic acid via EDC/NHS chemistry, whereas in the second strategy surface was passivated by the UV-hydrosilylation of 1,8-nonadiyne, and the alkyne-terminated ss-DNA was immobilised on 1,8-nonadiyne by a click' reaction with a diazide linker (see ESI†). | ||
We have shown previously that 1,8-nonadiyne can stabilise silicon electronically from forming oxide layer during electrooxidation.17 The resistance to oxidation has been attributed to the Si–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–R linkage, and the distal ethynyl moieties of the monolayer having an affinity for each other due to π–π stacking.6,19 How the surface will perform when DNA is attached is unknown, hence this surface chemistry will next be evaluated for fabricating selective DNA hybridization interfaces. Impedance spectra of the surface (SAM-6), prepared following strategy (b) of Scheme 1, were measured before and after hybridization with complementary and non-complementary DNA. Rct after hybridization with complementary DNA is 385 ± 53% higher as compared to the ss-DNA (see ESI†). As mentioned above, this observed increase in impedance is generally attributed to the electrostatic repulsion of anionic redox couple, [Fe(CN)6]3−/4−, with the negative charge on the surface from the additional phosphate backbone of the complementary DNA. Since a similar increase is also observed for non-complementary DNA (see ESI†), other factors apart from hybridisation of the DNA must be contributing. To investigate whether it is related to non-specific adsorption of target DNA on the sensing interface, electrodes exposed to both the complementary and non-complementary DNA were rinsed with ethanol. Rct was found to decrease with rinsing in both the cases indicating non-specific adsorption of DNA on the surface. Here it is worth mentioning that Rct for SAM-6 remains almost constant with consecutive potential cycling confirming the absence of surface oxides, hence 1,8-nonadiyne with ss-DNA attached to it can stabilise Si(111) electronically.
C–R linkage, and the distal ethynyl moieties of the monolayer having an affinity for each other due to π–π stacking.6,19 How the surface will perform when DNA is attached is unknown, hence this surface chemistry will next be evaluated for fabricating selective DNA hybridization interfaces. Impedance spectra of the surface (SAM-6), prepared following strategy (b) of Scheme 1, were measured before and after hybridization with complementary and non-complementary DNA. Rct after hybridization with complementary DNA is 385 ± 53% higher as compared to the ss-DNA (see ESI†). As mentioned above, this observed increase in impedance is generally attributed to the electrostatic repulsion of anionic redox couple, [Fe(CN)6]3−/4−, with the negative charge on the surface from the additional phosphate backbone of the complementary DNA. Since a similar increase is also observed for non-complementary DNA (see ESI†), other factors apart from hybridisation of the DNA must be contributing. To investigate whether it is related to non-specific adsorption of target DNA on the sensing interface, electrodes exposed to both the complementary and non-complementary DNA were rinsed with ethanol. Rct was found to decrease with rinsing in both the cases indicating non-specific adsorption of DNA on the surface. Here it is worth mentioning that Rct for SAM-6 remains almost constant with consecutive potential cycling confirming the absence of surface oxides, hence 1,8-nonadiyne with ss-DNA attached to it can stabilise Si(111) electronically.
If non-specific adsorption is responsible for the increase in impedance, the addition of bovine serum albumin (BSA) should provide significant improvement in differentiating different target DNA analytes. To test the hypothesis, ss-DNA-modified Si(111) (SAM-6) surfaces were incubated in a solution of 1% BSA + 0.05% SDS overnight before using it to detect target DNA (Fig. 1a). A large increase in Rct is observed upon the addition of complementary DNA (%ΔRct = 590 ± 38%) and is consistent with values reported previously.20 Slight changes are observed with the non-complementary DNA. This increase in impedance is similar to that of ss-DNA surface being incubated in only hybridisation buffer (52.1 ± 5%, data not given). At a neutral pH, BSA is negatively charged and provides a repulsive force to resist non-specific DNA binding. The results indicate that BSA acts as a good antifouling agent for the studied surfaces allowing the successful discrimination between target DNA analytes. The same electrode was further investigated to check its ability in discriminating between single-base mismatches. C-A and G-A mismatches display an increase in Rct by 147 ± 31% and 119 ± 28%, respectively, compared to the ss-DNA (Fig. 1b). The changes in impedance observed for the single-base mismatches are higher than those previously reported.21 However the sensing interface still has the ability to differentiate among the complementary, non-complementary and mismatched DNA.
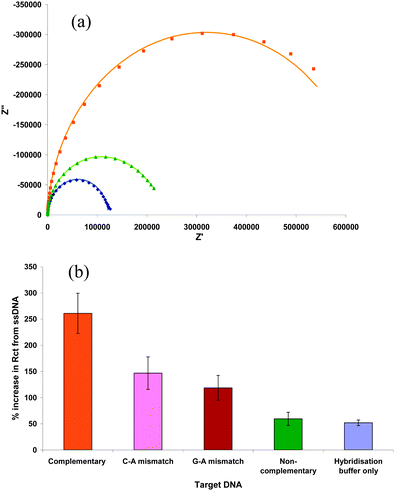 | ||
Fig. 1 (a) Nyquist plots for ( ) single stranded, ( ) single stranded, ( ) complementary, and ( ) complementary, and ( ) non-complementary DNA, and (b) comparison of the change in Rct for different target DNA analytes. Measurements were made after incubating in BSA. This figure shows that with BSA as antifouling agent, the modified surface [SAM-6, strategy (b) of Scheme 1] can be used successfully to detect DNA. All the Plots were measured in a solution of 5 mM [Fe(CN)6]−3/−4 + 0.1 M KCl at 0.2 V. ) non-complementary DNA, and (b) comparison of the change in Rct for different target DNA analytes. Measurements were made after incubating in BSA. This figure shows that with BSA as antifouling agent, the modified surface [SAM-6, strategy (b) of Scheme 1] can be used successfully to detect DNA. All the Plots were measured in a solution of 5 mM [Fe(CN)6]−3/−4 + 0.1 M KCl at 0.2 V. | ||
In summary, an interface prepared via UV-hydrosilylation of 1,8-nonadiyne on Si(111) has been develop for the detection of DNA. This sensor satisfy the basic criteria of being a robust and selective sensor, i.e., it stably immobilised DNA and resist the formation of insulating surface oxides during electrochemistry which changes the electronic properties of the interface. Using BSA to prevent non-specific adsorption of DNA, the recognition interface can successfully detect complementary, non-complementary and single-base pair mismatched DNA. Hence, we believe this is the first DNA modified surface suitable for the electrochemical detection of DNA hybridization on silicon electrodes.
We thanks the Australian Research Council's Discovery Projects Funding Scheme (DP1094564), Australian Research Council Centre of Excellence in Convergent Bio-Nano Science and Technology (project number CE140100036) and the University of New South Wales for funding.
Notes and references
- J. J. Gooding, Electroanalysis, 2002, 14, 1149 CrossRef CAS; O. I. Wilner and I. Willner, Chem. Rev., 2012, 112, 2528 CrossRef PubMed.
- A. Sassolas, B. D. Leca-Bouvier and L. J. Blum, Chem. Rev., 2008, 108, 109 CrossRef CAS PubMed.
- J. J. Gooding, Electroanalysis, 2008, 20, 573 CrossRef CAS.
- G. Liu, M. Chockalingham, S. M. Khor, A. L. Gui and J. J. Gooding, Electroanalysis, 2010, 22, 918 CrossRef CAS; Y.-C. Liu and R. L. McCreery, J. Am. Chem. Soc., 1995, 117, 11254 CrossRef.
- D. L. Allara, Biosens. Bioelectron., 1995, 10, 771 CrossRef CAS; A. Ulman, Chem. Rev., 1996, 96, 1533 CrossRef PubMed.
- S. Ciampi, M. James, P. Michaels and J. J. Gooding, Langmuir, 2011, 27, 6940 CrossRef CAS PubMed; M. R. Linford, P. Fenter, P. M. Eisenberger and C. E. D. Chidsey, J. Am. Chem. Soc., 1995, 117, 3145 CrossRef.
- J. K. Kariuki and M. T. McDermott, Langmuir, 2001, 17, 5947 CrossRef CAS.
- J. Pinson and F. Podvorica, Chem. Soc. Rev., 2005, 34, 429 RSC.
- T. Strother, W. Cai, X. S. Zhao, R. J. Hamers and L. M. Smith, J. Am. Chem. Soc., 2000, 122, 1205 CrossRef CAS.
- J. J. Gooding and S. Ciampi, Chem. Soc. Rev., 2011, 40, 2704 RSC.
- F. Decker, F. Cattaruzza, C. Coluzza, A. Flamini, A. G. Marrani, R. Zanoni and E. A. Dalchiele, J. Phys. Chem. B, 2006, 110, 7374 CrossRef CAS PubMed.
- R. L. McCreery, Chem. Mater., 2004, 16, 4477 CrossRef CAS.
- R. J. Hamers, Annu. Rev. Anal. Chem., 2008, 1, 707 CrossRef CAS PubMed.
- H. B. Yin, T. Brown, J. S. Wilkinson, R. W. Eason and T. Melvin, Nucleic Acids Res., 2004, 32(14), e118 CrossRef CAS PubMed.
- M. Steichen and C. Buess-Herman, Electrochem. Commun., 2005, 7, 416 CrossRef CAS PubMed; M. Manesse, V. Stambouli, R. Boukherroub and S. Szunerits, Analyst, 2008, 133, 1097 RSC.
- P. Allongue, C. H. de Villeneuve and J. Pinson, Electrochim. Acta, 2000, 45, 3241 CrossRef CAS.
- S. Ciampi, P. K. Eggers, G. L. Saux, M. James, J. B. Harper and J. J. Gooding, Langmuir, 2009, 25, 2530 CrossRef CAS PubMed; S. Ciampi, T. Böcking, K. A. Kilian, M. James, J. B. Harper and J. J. Gooding, Langmuir, 2007, 23, 9320 CrossRef PubMed.
- C. Steinem, A. Janshoff, V. S. Y. Lin, N. H. Völcker and M. R. Ghadiri, Tetrahedron, 2004, 60, 11259 CrossRef CAS PubMed.
- S. R. Puniredd, O. Assad and H. Haick, J. Am. Chem. Soc., 2008, 130, 13727 CrossRef CAS PubMed; L. Scheres, M. Giesbers and H. Zuilhof, Langmuir, 2010, 26, 4790 CrossRef PubMed.
- S. Pan and L. Rothberg, Langmuir, 2005, 21, 1022 CrossRef CAS PubMed.
- X. Li, J. S. Lee and H.-B. Kraatz, Anal. Chem., 2006, 78, 6096 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental, cyclic voltammograms and XPS data. See DOI: 10.1039/c4cc03418j |
| This journal is © The Royal Society of Chemistry 2014 |
